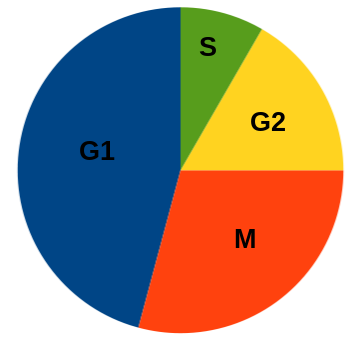
Idealized cell cycle, giving roughly proportional times. The times of cell cycle phases will differ with species and cell type.
1. The cell cycle separates gene
expression, DNA replication and cell division into discrete
phases. Be able to describe the main events in G1, S, G2 and
M.
2. The cell spends most of its time in interphase, during
which chromosomes are uncoiled, allowing gene expression to
occur. Know the main components and functions of the
interphase nucleus.
3. Be able to describe the main
events in mitosis, and be able to identify which stage of
mitosis a particular cell is in:
a. Prophase - condensation of chromosomes; disassembly of nuclear envelope into vesicles, attachment of spindle fibers.
b. Metaphase - Centrosomes pull spindle fibers taut, causing chromosomes to migrate to center of cell.
c. Anaphase - poleward movement of chromosomes through shortening of kinetochore spindles, after which poles are pushed apart by polar spindles.
d. Telophase - laminar vesicles reassemble into nuclear envelope; chromosomes decondense; cytokinesis
| In the eukaryotic cell, the cell cycle
divides the activities of gene expression, chromosome
replication and cell division. Under the microscope, somatic
cells seem to spend most of their time doing nothing. These
long periods of inactivity are punctuated by relatively
short periods where DNA synthesis and mitosis take place. Idealized cell cycle, giving roughly proportional times. The times of cell cycle phases will differ with species and cell type. |
Typical Eukaryotic Cell
Cycle
 |
| The following experiment defined the phases
of the eukaryotic cell cycle: Cells in culture can be made to grow synchronously, which means that all the cells divide at the same time. To follow events in the cell cycle, you can feed the cells with radioactive nucleotides at different times, and measure the radioactivity remaining in cells after washing. Incorporation of radioactive nucleotides indicates that DNA synthesis was occurring at the time of sampling. There is only a narrow time slot in which cells take up tritiated thymidine. This is the S phase, for DNA synthesis. |
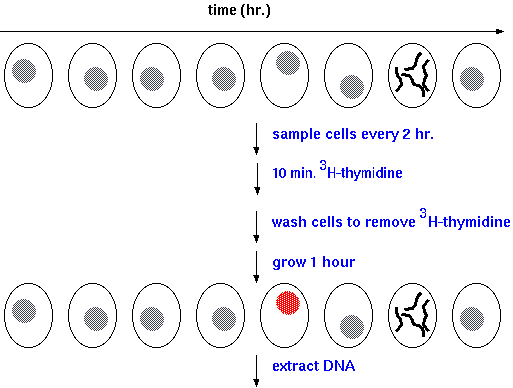 |
The phases of the cell cycle are organized by the two major cell
changes: DNA replication and cell division. The other two phases
are "gap" phases between those two activities.
1) G1 ("gap in DNA synthesis") - most GENE EXPRESSION occurs here. In non-dividing or terminally differentiated cells, G1 persists indefinitely, and is referred to as G 0. Chromatin is dispersed in nucleus.
2) S - (DNA synthesis); The cell requires a discrete signal to begin a round of DNA replication. eg. if you fuse a cell in S phase with a cell in G 1, the nucleus from G 1 will begin DNA synthesis.
3) G2 ("second gap in DNA synthesis") The nucleus is reorganizing in preparation for mitosis. Some chromatin condensation occurs here, but not enough to be visible under light microscopy.
4) M - ("mitosis or meiosis") - chromosomes partitioned between two daughter cells.
Interphase, which includes G1 or G0, S and G2, is where the cell spends most of its time. Interphase is where most gene expression occurs. Put another way, most of what the cell does, other than dividing, occurs during interphase.
|
|
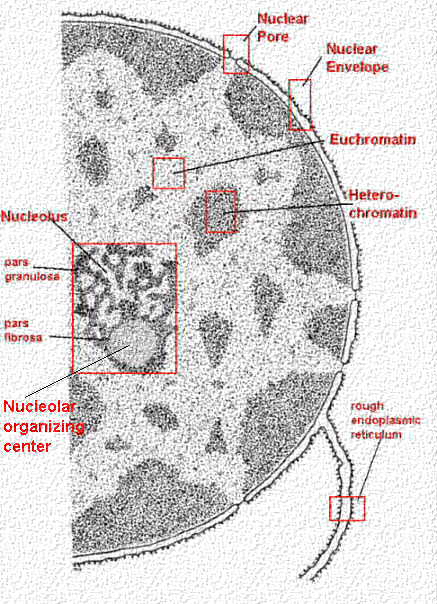 |
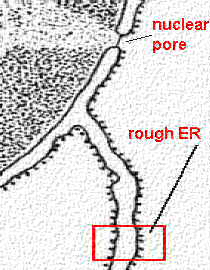
The envelope is actually two membranes: the inner membrane has attachment sites for chromosomes, and the outer membrane helps define the shape of the nucleus. The space between these two membranes is called the perinuclear space.
The traffic of different molecules in and out of the nucleus is
controlled by nuclear pores which are protein
complexes of their own.
| VIDEO: Into the Nucleus - animation of import into nucleus via nuclear pore http://youtu.be/UyhqLpjicZg | The nuclear pore complex consists of
stringlike fibrils that project into the cytoplasm, a
central meshwork, and a nuclear basket on the inside. 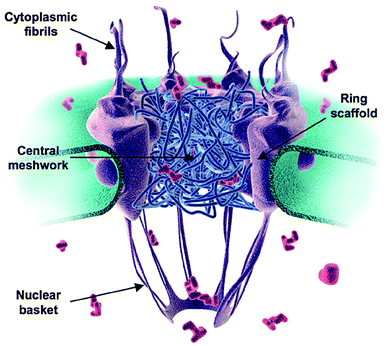 Displayed from http://pubs.rsc.org/services/images/RSCpubs.ePlatform |
Generally, molecules can go in or out of the nucleus - not both. RNA molecules made inside the nucleus are transported out for translation by ribosomes. Proteins made outside the nucleus are transported in for use inside.
What are some of the proteins that get transported into the nucleus?
The nucleus may have evolved from the endoplasmic reticulumThe nucleus is the main difference between prokaryotic (pre-nucleus) and eukaryotic (true nucleus) cells. So how did this nucleus come to be? If we look closer at the image from above, we can see that the nuclear envelope seems to be continuous with the rough endoplasmic reticulum (RER).If we consider the nuclear envelope as a differentiated form of the RER, there would have been some clear advantages to cells which formed a nucleus. RNA transcripts that exit the nucleus come into immediate contact with ribosomes for translation. Proteins translated that need to go back to the nucleus don't have far to go. In this model, the formation of a nucleus would have been preceded by the formation of a rough (ribosome-prevalent) endoplasmic reticulum in the first place. |
 |
| The distinction between
mitosis and meiosis Mitosis - nuclear division leading to equal sets of chromosomes in the daughter nuclei. Characteristic of somatic cells. Meiosis - nuclear division leading to a reduction in chromosome number of the parent cell to produce haploid gametes. |
The mechanism of mitosis
During interphase, the chromosomes need to be loose so that gene expression can take place. But when it comes time for the cell to divide, loosely coiled chromosome spaghetti isn't easy to separate. So, the chromosomes have to get a lot smaller and denser to divide in an orderly fashion.
When the chromosomes have coiled enough that they retain stains (and we can see them), we call that prophase. The nuclear envelope is also dissasembled during prophase. In metaphase, we see the chromosomes attached to spindle fibres and lined up at the centre of the cell. During anaphase, the "tug-of-war" between the chromosomes and centrosomes resolves, and we see the chromosomes being dragged to opposite ends of the cell. Finally, in telophase, the nuclear envelope reassembles, the chromosomes decondense, and the nucleus reorganizes itself, allowing normal cellular functions to resume.
| It is important to remember that interphase
is defined by what we can see under the microscope with
conventional staining techniques. While it may look
like nothing is happening, we have already seen that the
term "Interphase" includes G1, S and G2 phases of the cell
cycle. |
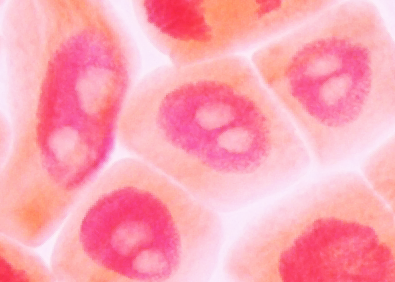 Onion
root tip cells in interphase, stained with aceto
carmine. Nuclei stain red, nucleoli appear white.
Onion
root tip cells in interphase, stained with aceto
carmine. Nuclei stain red, nucleoli appear white. Image from Yaping Wang and Brian Fristensky, University of Manitoba. |
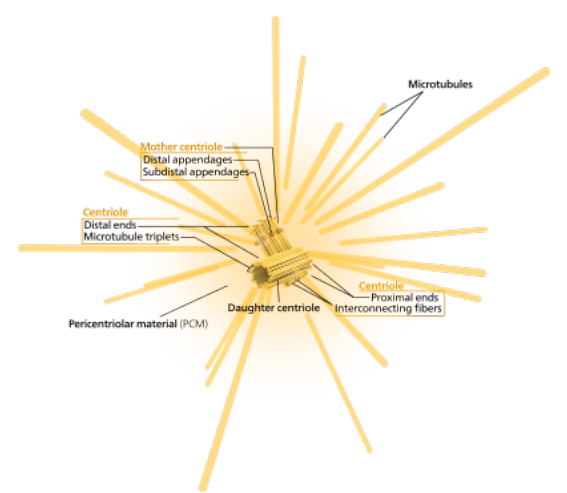
The role of centrosomes in the cell cycle are illustrated in the figure. (from http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=cooper.figgrp.2472 ) Prior to the visible onset of prophase, the following steps occur:
|
Centrosomes are the site of spindle fiber
organization, and thus provide a polarity to the dividing
cell. They also serve as "basal bodies" in cells
with flagellae. From https://en.wikipedia.org/wiki/Centrosome |
 |
| Spindle fibers are microtubules which originate at the centrosomes. Microtubules are tubes formed from polymers of α- and β-tubulin heterodimers. As illustrated above, bundles of microtubules also form the centrosomes. Centrosomes are also referred to as "microtubule organizing centers" (MTOC). | 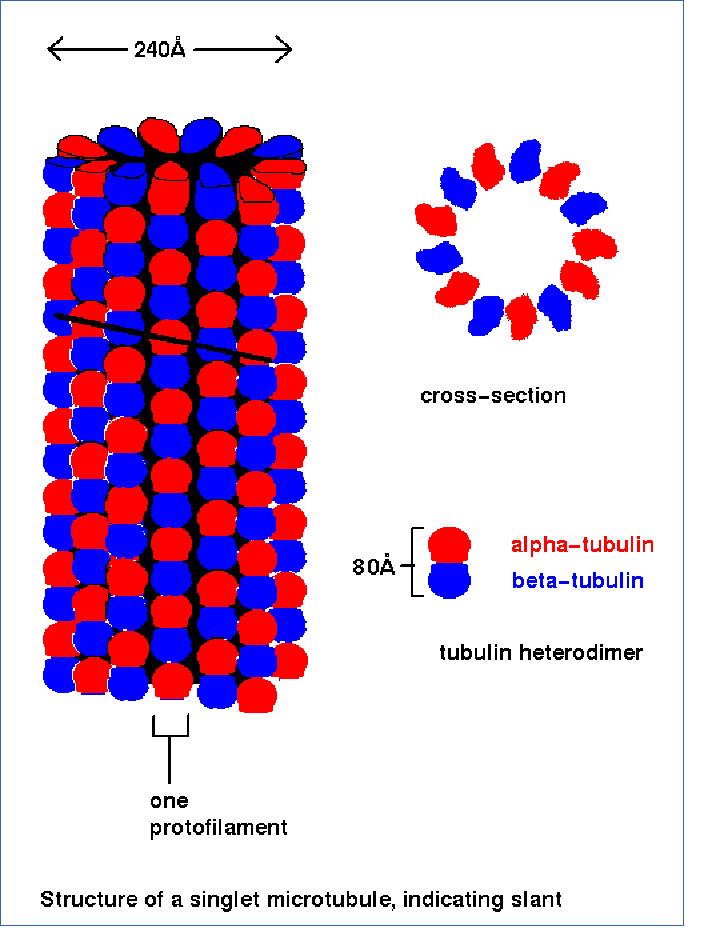 |
| NOTE:
Remember the cell cycle experiment. Long before we see the
chromosomes condensing at prophase, the chromosomes have
already replicated! |
| Prophase in onion
root tips with aceto-carmine staining. A - C show the
progression of chromosome condensation. Note that as
chromosomes condense, they become thicker and less
diffuse. Images from Yaping Wang and Brian Fristensky, University of Manitoba. |
A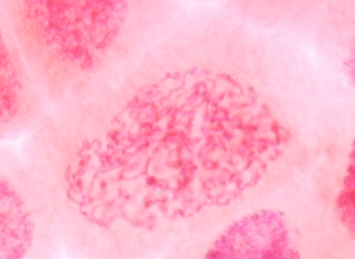 |
B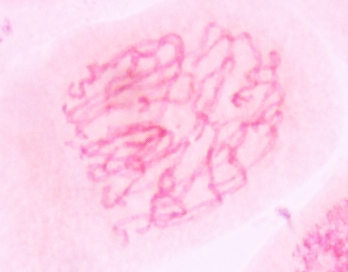 |
C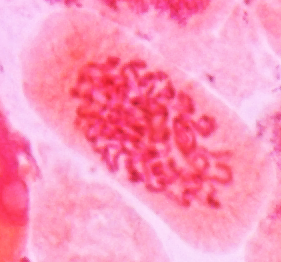 |
2. Switch in microtubule stability accompanies formation of spindle fibers.Spindle fibers extend randomly in all directions. The length of the fibers varies due to polymerization and depolymerization of the microtubule from tubulin dimers. Polymerization occurs at both ends, but is relatively slow at the (-) end, which is anchored at the centrosome. The (+) end grows more rapidly, as it extends into the cytoplasm. However, when a microtubule encounters the kinetochore at the centromere of a chromosome, it's (+) end is stabilized by the kinetochore. The kinetochore is a protein matrix, consisting of proteins that attach to centromeric DNA sequences (inner layer) and proteins that attach to the spindle fibers (outer layer).The kinetochore is a protein/DNA complex which forms at the centromere. It is the site of attachment of kinetochore microtubules to the mitotic chromosome. |
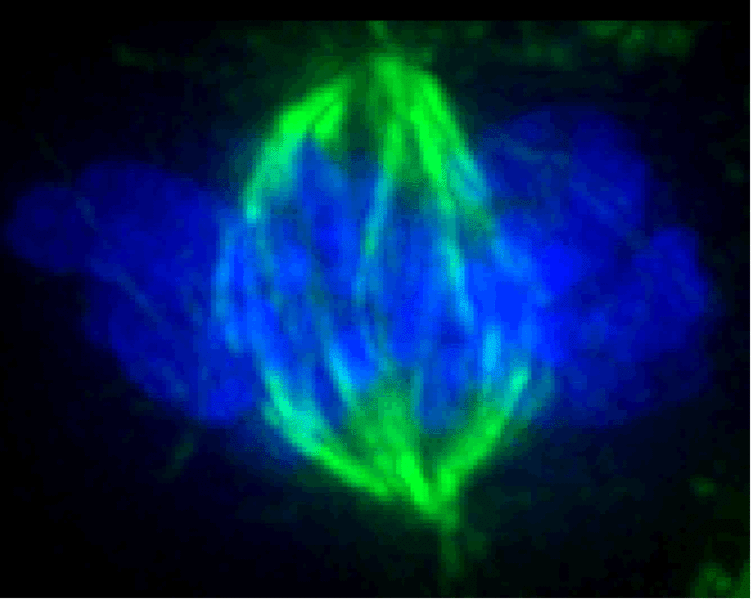 Fluroescently-tagged spindle fibers are shown in green. DNA in chromosomes is counterstained in blue flurorscence. http://en.wikipedia.org/wiki/Image:Mitotic_spindle_color_micrograph.gif |
 Displayed from https://www.nature.com/scitable/content/18070/10.1038_nrm2310-f2_large_2.jpg |
In summary, prophase is characterized by:
| In metaphase the chromosomes are
fully condensed and arranged in a plane equidistant from
the poles of the spindle. This is the highest level of
coiling, and the chromosomes are shorter and thicker
than any other stage and therefore ideal for cytogenetic
study. There is no longer much relational coiling
present, and the chromatids lie side by side, not
twisted around each other. Experimental evidence indicates that centrosomes appear to pull the spindle fibers taut, in a kind of "tug of war". Thus, the chromosomes migrate toward equator of the cell and become aligned. VIDEO (12.7 Mb) PLNT3140-CongressionOfChromosomes.webm |
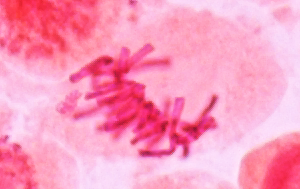 Metaphase
in
onion root tips. |
In summary, metaphase is characterized by:
Anaphase is the shortest phase of mitosis. The beginning of
anaphase is observed microscopically when the synapsed
sister-chromatids separate simultaneously, and begin migrating to
the opposite cellular poles. During anaphase, longer chromosomes
often appear V-shaped or J-shaped.
Early anaphase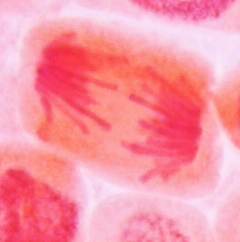 |
Late Anaphase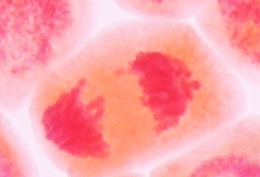 |
| Images
from
Yaping Wang and Brian Fristensky, University of
Manitoba. |
|
polar fibers - spindles
interdigitated with spindles from opposite poles
At the end of anaphase, the spindle fibres converge at the pole and the chromosomes are jammed together. Polymerization of + ends of polar fibers push the poles apart, bring the chromosomes with them.
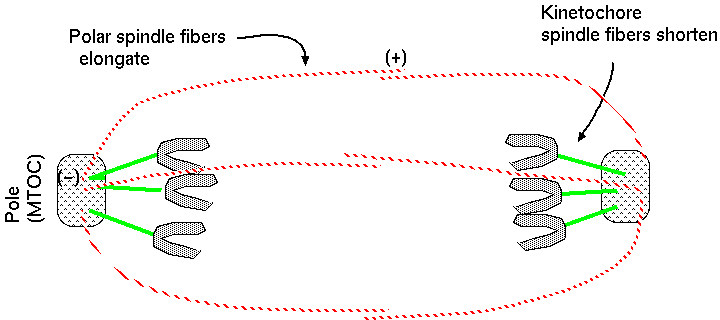
In summary, anaphase is characterized by:
| Telophase starts when chromosomes reach
opposite poles and form a dense chromatid ball. It is the
termination of mitosis and the most difficult stage to study
cytogenetically because the chromsomes are crammed into a
small space, which will reconstitute into an interphase
nucleus. During telophase, the nuclear envelope is reconstituted from vesicles leftover from the parent nucleus, and chromsomes begin to decondense. Nucleoli, the nuclear matrix and the nuclear membrane reform, and the spindle fibers disassemble. Telophase restores the daughter cells to the interphase state. |
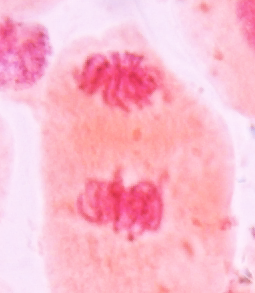 |
 Displayed from
http://upload.wikimedia.org/wikipedia/commons/8/8a/Nuclear_envelope_breakdown_and_reassembly_in_mitosis.jpg Displayed from
http://upload.wikimedia.org/wikipedia/commons/8/8a/Nuclear_envelope_breakdown_and_reassembly_in_mitosis.jpg |
Telophase restores the daughter cells to the interphase state. Envelope vesicles first enclose each individual chromosome, after which vesicles fuse to form a single genetically complete nucleus at each pole. Nucleoli and the nuclear membrane reforms and the spindle fibres disappear. Chromatin decondenses, and the nuclear matrix re-assembles.
The division of the cytoplasm and its organelles between daughter cells is called cytokinesis, which begins during late telophase at the equatorial plate. In plants, cytokinesis takes places by the formation of the cell plate. In animals, cytokinesis works by formation of a cleavage furrow, pinching off the cell much like a drawstring. Generally, cytokinesis divides the parent cell equally between the two daughter cells. Sometimes, no cytokinesis occurs after nuclear division, resulting in a binucleate cell. Additionally, division can be asymmetrical and give rise to daughter cells of different shapes and sizes.
In summary, telophase is characterized by: