Lecture 8, part 2 of 2
| PLNT4610/PLNT7690
Bioinformatics Lecture 8, part 2 of 2 |
The object concept can be stated as follows:
|
The fundamental
unit of Object-oriented databases (OODB) is the Class.
A class can
be thought of as a formula for an object.
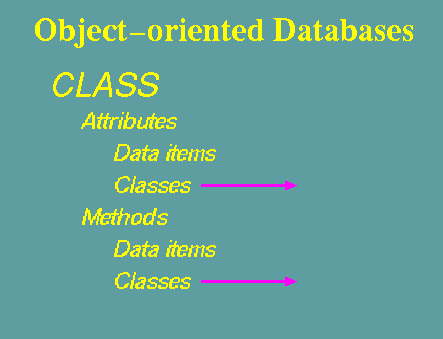
A class may have many attributes and methods.
An object is an instance of
a class
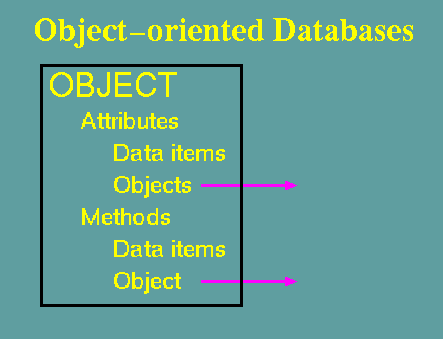
Objects, in OODBs, correspond to individual records in a relational database. There may therefore be many objects of a given class. A class is an abstraction. An object is a tangible thing.
pFF100 is an instance
of the class PLASMID. The class definition allows it to point to
two other classes, VECTOR and DNA_SAMPLE. DNA_SAMPLE might look
like this:
| CLASS | OBJECT |
DNA_SAMPLE
VECTOR | PLASMID |
SK99.pFF100
pFF100 |
In the DNA_SAMPLE
class, the first field points to either a VECTOR or a PLASMID.
In this case, the corresponding field in SK99.pFF100 points to
PLASMID pFF100. the EXPERIMENT field points to an object of the
type EXPERIMENT. the concentration field contains a floating
point number whose units are µg/ µl. The BOX field points to an
object of the type BOX, called "Pisum ESTs I".
| A database is a model of something in
the real world It is important to note that out of 4 fields in the DNA_SAMPLE class, three of them are relations. That is, three of them point to other classes. Only one contains data. This illustrates the point that relations between objects are often as important as the data themselves. The relations between objects give the data objects characteristics similarly to their real-world counterparts. . |
| CLASS | OBJECT |
PLASMID
VECTOR |
pFF100
pBluescriptIISK+ |
Here, the MAP_VIEWER and filename fields could be a template for a command that would launch a viewing program with a specific file. In the pFF100 object, the actual command that would be run is ' pFF100.gif'. This command would be passed to UNIX, launching the eog image viewer with the GIF file pFF100.gif.
Depending on the
software, it might even be possible to call a different viewing
program for different types of image files. For example, if a
plasmid called pMU1 had a map in Adobe PDF format, it would be
necessary to view its map using the Adobe Acrobat viewer eg.
'acroread pMU1.pdf'.
| CLASS | OBJECT |
VECTOR
PLASMID |
pBluescriptKSm13+
pI206KS |
Here, four plasmid
constructs were made using the pBluescriptKSm13+ vector, and
three DNA_SAMPLEs of this vector (not the plasmids) are listed.
One point to make here is that when a class is changed, not all objects in that class need to be modified. OODBs, and to some extent other types of databases, do not require that all objects contain data for all possible attributes defined in the class. This allows a 'grandfathering' of preexisting objects. For example, if the class Cell_Stock was modified by the addition of an attribute called 'Date', listing the date on which the stock was made, it would not be necessary to go back and insert a date into (potentially thousands) of existing Cell_Stock objects. Dates can be included in new Cell_Stock objects, as they are created.
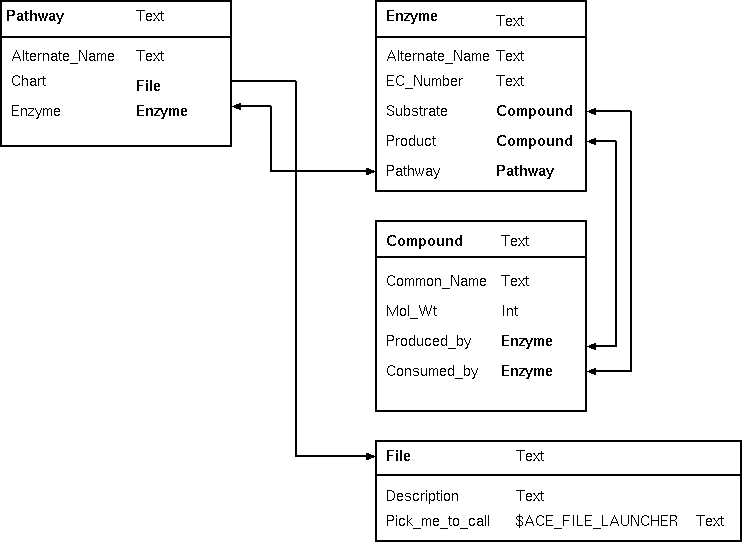
| The schema at right implements a biochemical
pathways, using the conventions of the ACeDB system. Each
pathway object points to one or more enzymes present
in that pathway, and each enzyme object points to one or
more pathways to which it belongs. |
 |
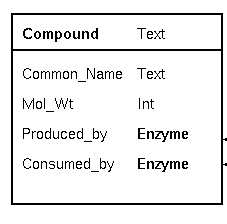
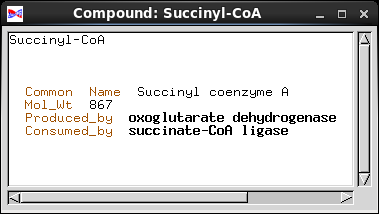
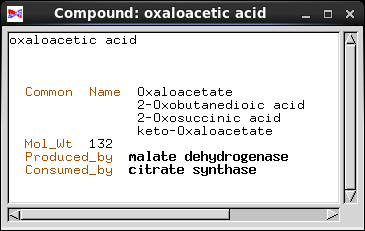
| Databases try to create a model of real-world
things as we understand them. To make this possible, it is
useful to give each field a label, which describes what each
piece of data is intended to represent. The label is a
convenience for the human user. Each field also has a data
type, which indicates the type of data used to represent
that piece of information. Common_Name is implemented as a Text field, a string of characters. Mol_Wt (molecular weight) is a number, so it is implemented as an integer. In a biochemical pathway, a compound can be a product of one enzyme, and a substrate for one enzyme. To represent these concepts, we have two fields, Produced_by and Consumed_by. Both point to objects of the Enzyme class. Note: Common_Name and Mol_Wt are examples of fields in which the information is contained in each object. Produced_by and Consumed_by are examples of fields which point to other objects. |
 |
 |
 |
| Pathway Demo You can try a database that implements this schema to emulate the TCA cycle, by typing 'pathace' at the Linux prompt. |
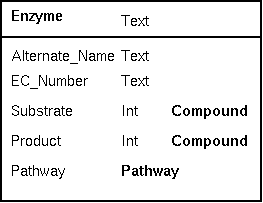
| One
of the best tests of a well-thought out a database
occurs when you decide that the schema needs to be
modified to add new concepts. For example, the existing
Enzyme class could be extended to incorporate the concept
of stoichiometry by adding coefficients to each compound
linked-to in the enzyme class. In the example at right, an
integer (Int) tells the number of molecules of a substrate
or product consumed or produced. Adding these fields doesn't require any changes in the other data objects. If your classes are well-designed, a change in one class will not break other classes. |
 |
1. The database is a model of a biological or experimental system. Make it as close to the real system as possible.
2. Keep each class simple. The fewer fields, the better.
3. Do not duplicate the same piece of information in more than one object.
4. Wherever practical, avoid free text. Use links or enumerated choices.
Alvare, G., Roche-Lima, A. & Fristensky, B. BioLegato: a programmable, object-oriented graphic user interface. BMC Bioinformatics 24, 316 (2023). https://doi.org/10.1186/s12859-023-05436-4
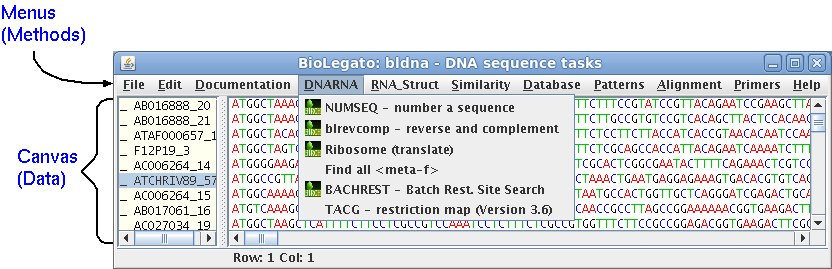
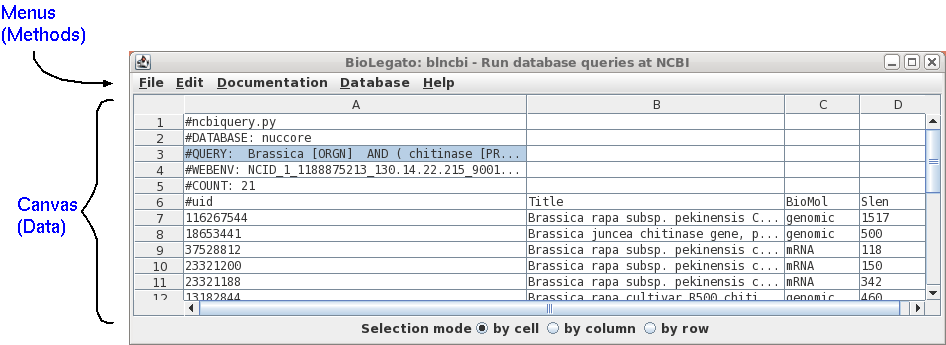
BioLegato is a fundamental rethinking about application programs. It takes as its premise the idea that objects are an intuitive way to combine information and the methods that work with that information. If the objects are structured like things that the end user is already familiar with, the fact that the user already understands the relationships between objects, and what they are expected to do, makes it easier to use the software.| blgeneric is a BioLegato application that launches BioLegato without any menus or canvas. This is mainly for demonstration purposes, to illustrate the fact that almost all functionality of BioLegato is programmable. In the terminology of Object-Oriented programming, think of BioLegato as an abstract class that is extended to create real classes. So in a way blgeneric is like instantiating an abstract class. To launch type 'blgeneric'. |  |
The Tree of Life is a
taxonomic database edited by David R. Maddison at the University
of Arizona.
Its main structure is a hierarchy of web pages, whose root is at
the kingdom level. Hypertext links allow a user to traverse the
phylogenetic tree from one level to another (eg. phylum, order,
class, family, genus, species). At each node, specialized data
of almost any kind may be found, from images to text documents,
or even links to other web sites.
|
DEMO: Descend Tree of life as follows: root
Organisms with nucleated
cells (Eukaryota)
Animals (Metazoa)
Bilateria
Deuterostomia
|
|
DEMO: Descend Tree of life as follows: root
Eukaryota
Animals (Metazoa)
Bilateria
Deuterostomia
|
Although it may seem like a subtle difference, the Tree of Life IS a collection of web pages, whereas the web pages visited at NCBI are generated on the fly from the NCBI database. The web pages seen at NCBI are therefore a view of the data.
It should be pointed
out that each approach has advantages and disadvantages. The
NCBI web site is formal and structured, but primarily serves to
encode a taxonomic structure, with links to databse items such
as sequences or literature references. The Tree of Life is rich,
with images, articles, and other information, limited only by
the creativity of the contributers.
 Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada |
| PLNT4610/PLNT7690
Bioinformatics Lecture 8, part 2 of 2 |