
 |
BIRCH - Adding binaries, scripts and
libraries |
| A point of emphasis: BioLegato is a program
that runs other programs. Put another way, just about any program on the
system, any program at all, can be launched from
BioLegato. You can launch a script or a binary.
The script could launch other scripts or binaries. If your
output is in the form of a table, you could launch a
spreadsheet. If the output is sequence, you can launch
another BioLegato window! You really have to use BioLegato
for awhile to appreciate how powerful this capability is. This section provides the information you need to add programs to BioLegato at your site. You will discover that actually, the hard part about adding a new program is first figuring out how to run that program at the command line. Subsequently, creating a BioLegato menu is straightforward. There are three critical things to know:
|
| bl_instance |
script |
menu directories |
| birch - launcher for BIRCH
programs |
$birch/script/birch |
$birch/dat/birch $birch/local/dat/birch |
| bldna - DNA sequence
analysis |
$birch/script/bldna | $birch/dat/bldna $birch/local/dat/bldna |
| blprotein - protein
sequence analysis |
$birch/script/blprotein | $birch/dat/blprotein $birch/local/dat/blprotein |
| blmarker - molecular
markers |
$birch/script/blmarker | $birch/dat/blmarker $birch/local/dat/blmarker |
| bltree - phylogenetic trees |
$birch/script/bltree | $birch/dat/bltree $birch/local/dat/bltree |
| Example of a BioLegato folder -
bldna |
| $BIRCH/dat/bldna bldna.properties
PCD
Alignment
Database DNARNA AssembleContigs.blmenu
.......Bachrest.blmenu BLrevcomp.blmenu Findall.blmenu NUMSEQ.blmenu pcd_order Ribosome.blmenu TACGrest.blmenu Similarity pcd_order |
| pcd_order file |
Appearance of menus, left
to right, in BioLegato |
| File Edit Documentation DNARNA RNA_Struct Similarity Database Patterns Alignment Primers Phylogeny Help |
 |
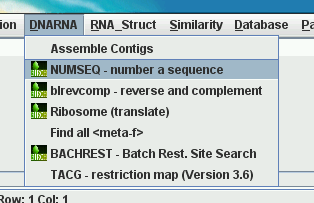
| pcd_order file |
Appearance of DNARNA menu,
top to bottom |
AssembleContigs.blmenu |
 |
| Before you start - You really need to have a good idea of how to
run your program at the command line before you
try to make a BioLegato menu item to run it. Read the
manual pages or other documentation. Try it out. It will
save you a lot of time and frustration. |
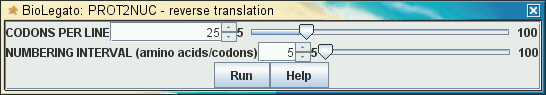
name "PROT2NUC - reverse translation"
icon "$BIRCH/public_html/images/birch_black_ico.png"
tip "use ambiguity symbols to represent all possible DNA sequences encoding each amino acid in a protein"
var "linlen"
type number
label "CODONS PER LINE"
min 5
max 100
default 25
var "in1"
type tempfile
direction in
format flat
save true
var "group"
type number
label "NUMBERING INTERVAL (amino acids/codons)"
min 5
max 100
default 5
panel
var "Run"
type button
label "Run"
shell "sed ""s/[#%]/>/"" <%in1% | prot2nuc -l%LINLEN% -g%GROUP% > %in1%.out; ($BL_TextEditor %in1%.out; rm %in1%*)&"
close true
var "Help"
type button
label "Help"
shell "$BIRCH/script/chooseviewer.py $BIRCH/doc/xylem/prot2nuc.txt"
close false
| The menu generated by the
above code looks like this: |
 |
| Note on
BioLegato HELP buttons The example above shows the syntax for specifying a help file to be opened in when the HELP button is pressed in a PCD item menu. The general formula is: var "Help" type button label "Help" shell "program filename" close false where filename is the file to open and program is the program to open the file. filename can also be a URL. program is usually gde_help_viewer.csh, which chooses the viewer based on the file extension. |
shell "sed ""s/[#%]/>/"" <%in1% >%in1%.test; $BL_TextEditor %in1%.test; cat %in1%.test | prot2nuc -l%LINLEN% -g%GROUP% > %in1%.out; ($BL_TextEditor %in1%.out; rm %in1%*)&"
will
let you verify that the sed command worked corrrectly before
prot2nuc is run.