Databases
and
Web Services
As we will
see today, a major trend in bioinformatics is the proliferation of
web services. Web services are the logical next step. Whereas
databases decentralize the storage and organization of
information, web services offload many computing tasks to other
systems. The major developments in web services for bioinformatics
are an outgrowth of the open computing concept, in that services
are developed using open source software, and made available to
the research world at no charge.
For today's lecture: Don't just think about
how you might use web data and web services. Think about
how you might
contribute knowledge from your specialized field, in the
form of data and web services, to the growing "semantic
web".
|
1.
Client/Server interfaces
To simplify access of
data from remote locations, client/server protocols are used. The
- Client - a
program that runs on a local machine, processing user
requests. The client "talks to" the a server program across
the Internet, sending instructions for a transaction, and
retrieving the results of that transaction, to be displayed
locally.
- Server
- a program that retrieves the requested data from a database,
and sends them back to the client.
a.
FTP - File Transfer Protocol
Email is not the best way
to move files across the network, if for no other reason than the
fact that it requires human intervention at both ends.
It might seem
obvious, but the ability to download or upload files across a
network is important, because it's often more useful to have
locally-installed copies of databases. Local copies are
useful for projects in which rapid retrieval of large numbers of
sequences are important, such as creating database subsets. FTP is
a special case of the more general Client/Server model.
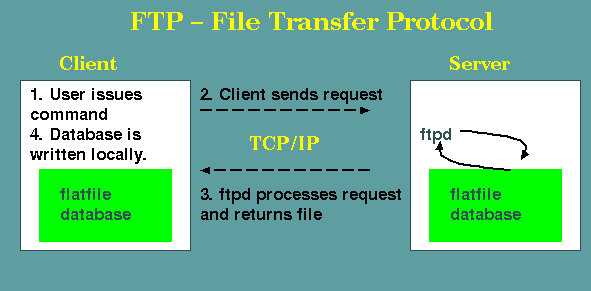
FTP programs that use Secure Shell (ssh) protocols
for encrypted file transfer:
Unix/Linux/Mac
sftp
- command line program
Windows, Mac, Linux
download - move files from a remote machine to your local
machine
upload - move files from your local machine to a remote
machine
Why have local copies of databases?
- Processing large numbers of transactions most
efficient on local filesystems
Remote database servers may be specialized to process
transactions one at a time. Local programs may allow batch
requests to local copies of database files.
- Many locally-installed programs can read a single
local flat-file database
Remote databases are typically managed through a single database
management system, whose files are unreadable by other
programs. Local flat-file databases can be read by any number of
programs
b. Interactive client/server programs
FTP is one special case
of the more general client/server model. A more typical case is
the NCBI BLAST+.
These are the standalone BLAST programs (including blastp, blastn,
tblastn, blastx and tblastx) that can run on any computer. By
default, the BLAST+ programs search local copies of NCBI
databases. However, if run with the -remote command line
option, they send the query to the NCBI, and the results return to
your local machine, as if you had run the search locally.
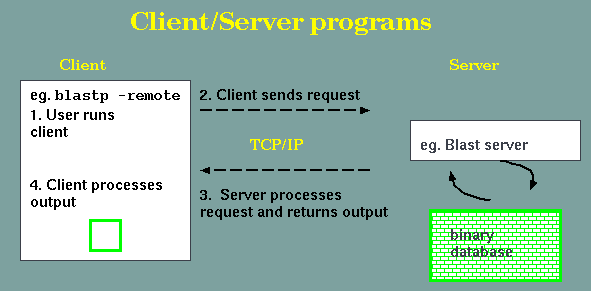
BLAST+ example
The following command
will search for a sequence in the NCBI GenBank non-redundant
(nr) protein database:
blastp
-remote -query PEADRRB.pro.fsa -db nr -out PEADRRB.blastp
This command tells
blastp to send a sequence to the NCBI Blast server, and run the
search using blastp to search the non-redundant protein
database. At the server end, the Blast server runs the search
and sends the data back to the client, which writes the output
stream to a file. Transactions between client and server are
carried out using the common internet protocol TCP/IP.
Transactions can only occur through remote server
In the client/server model the only way to send or
receive data to or from the database is with clients specifically
written for the particular server program that talks to the
database. This is good, in terms of system reliability, because
potentially, databases that are updated by user transactions could
conflict, which might result in a corrupt database. On the other
hand, the requirement for going through a specific server program
may limit the kind of things you can do with the database
Tasks can be strategically divided between
Client and Server
The Client/Server model provides an opportunity to offload some
tasks to the client. For example, most of the work of the user
interface is best done at the Client end. In particular, rendering
of graphics would be slow if done at the server and then
transferred to the Client.
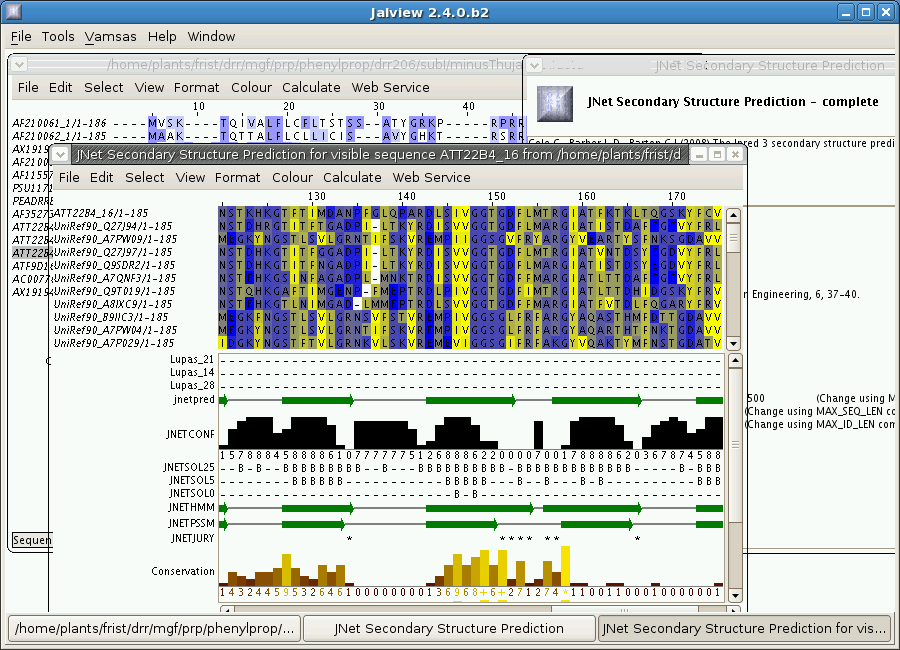
Example: Jalview multiple alignment
viewer
Jalview [http://www.jalview.org/]
is a Java program that runs on the user's computer. Its main
onboard functions are for visualizing multiple sequence
alignments. However, Jalview extends its functionality by running
web services. These services include:
- retrieval or
sequences and 3D protein structures
- multiple sequence
alignment
- protein secondary
structure prediction
- visualization of
protein 3D structures
In the example below, a
secondary structure prediction was done by the JNet service.
Secondary structure results are displayed below the sequence
alignment. For example, α helices are shown as red tubes, and β
sheets as green arrows.
2.
Web interfaces
Web interfaces to remote
databases are often easy to implement, and are easy to use. They
are easy to implement because minimal software development needs
to be done at the client end. The client is simply the Web
browser. All the work is done at the server end. The trick is to
get HTTP requests translated into a form the database software can
understand, and to convert output from the database program
into HTML and graphics.
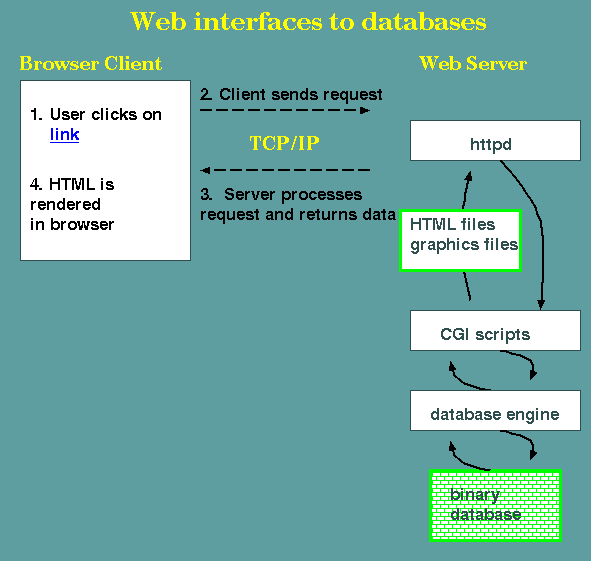
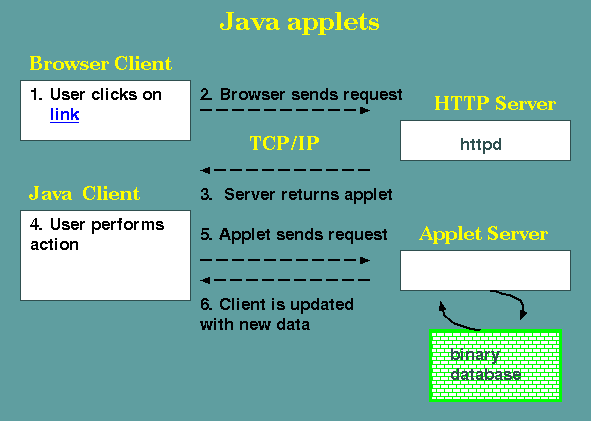
The figure shows that
as with all Web pages, the HTTP daemon httpd receives an HTTP
request, which is processed by a CGI script. A CGI script
contains instructions for running programs at the server end. In
this case, the CGI script would run programs that call the
database software, asking for the requested data. The data is
returned to the script, which runs further programs to create
HTML and graphics. The HTML and graphics are sent to httpd,
which passes them on to the remote Web client.
Example of a link that
calls CGI scripts:
https://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=AA960716&report=GenBank
This URL passes
commands to the Entrez server at NCBI to retrieve from the
GenBank nucleotide database the entry whose ACCESSION number is
AA960716.
The organization of
tasks makes it possible to use almost any database software at
the server end, without modification. The CGI scripts and
associated programs, along with httpd, act as "middleware",
between client and server. Even if, at some later time, the
structure of the database changes, or a different database
program is used, only a small amount of code needs to be
rewritten. The user can be given exactly the same view of the
data, regardless of what changes have been made to the database
itself.
There are several
important limitations to Web interfaces. First, web pages
display one page at a time. Every time a web page is updated,
the whole page must be redrawn. Updating a page is often
accompanied by additional transactions between client and
server, which may result in further delays. Furthermore, Web
browsers are usually oriented to a single window. Users move
from one window to the next, rather than having multiple windows
displayed simultaneously. Each browser window carries a
substantial deal of processing overhead.
3.
Java
Web Clients
a.
The Java language - "Write Once, Run Anywhere"
Java is an object-oriented
programming language designed at Sun
Microsystems, and now supported by
Oracle . It is popular for many reasons, one of them being
that it was specifically designed to be platform-independent.
Platform independence is accomplished in two ways. First, the
specification of the language has no platform-specific
dependencies. That is, there are no calls to programs or libraries
specific to any particular operating system. For example, Java
contains its own procedures for drawing windows, rather than
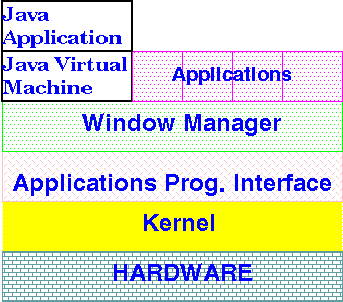
relying on system-specific libraries. Secondly, Java applications
are compiled (translated) into machine code that runs in the Java
Virtual machine (JVM). JVM maps Java instructions to actual
machine instructions. JVM can be thought of as an emulated
computer - a computer that runs as software rather than hardware.
Therefore, JVM needs to be adapted for each computer system
on which Java will run. Since JVM is now available for essentially
all computer platforms, Java programs can run, unmodified, on all
platforms.
On Linux systems,
for example, Java applications might be displayed by the Xfce
window manager, and some X11 calls might be issued by the JVM to
create windows. The kernel, ultimately, executes all
instructions emulated in the JVM.
b.
Java applets
The Java Virtual Machine,
JVM, is surprisingly small. Therefore, the major Web browsers
include a JVM that allows them to run Java "applets". Applets are
Java applications that are downloaded from a server at runtime,
but run in a local JVM, by the Web browser. As a security measure,
the JVM is implemented as a "sandbox", that is, a
virtual machine that can not read or write anywhere except in a
protected area of memory. No disk files can be read or written,
and no instructions can be executed outside of the sandbox. In
contrast, normal Java applications, run from an user's account,
can execute with the same read and write capabilitiies of any
other program.
Example: The 3D structure of the
nucleosome can also be viewed using Java applets at The Protein Data Bank
http://www.pdb.org/pdb/explore/explore.do?structureId=2CV5
Advantages
- Java applets run as independent windows, or
within the browser
Web browsers tend to move from one page to another, defeating
the purpose of having multiple windows. Applets can run in
multiple windows for different types of data, or different
procedures.
- Java applets can implement more sophisticated
user interfaces than are possible through HTML
HTML only has very limited capabilities for user input and
display of data. Applets can work on the data locally, in real
time, with any type of control desired eg. sliders, scroll bars.
- Java applets completely platform independent.
"Write once.
Run anywhere".
For most purposes, the applet can't not run, regardless of the
computer system at the client end. Only one version needs to be
written, rather than many different versions for different
platform. Thus, a Java program will typically run an Windows,
Mac, Unix, Linux, and probably your cellular phone.
- Java applets are not permanently installed at
client end.
Since the Java applet is newly-downloaded at runtime, the most
recent version of the applet will always be running at the
client end.






 Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada
Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada