DRR206 IS ACTIVATED WITH RACE-SPECIFICITY BY BACTERIA
AND FUNGI
The DRR206 gene family encodes a protein
of approx. 20 kDa with no significant similarity to any known protein.
garnier plot of gde22861_2.seq, 184 aa; DCH = 0, DCS = 0
>DRR206C
. 10 . 20 . 30 . 40 . 50 . 60
MGSKLPVLFVFVMLFALSSAIPNKRKPYKPCKNLVLYFHDILYNGKNAANATSAIVAAPE
helix HHHHHHHHH HHHHH HHH
sheet EEEEEE E EEEEEEEEEEE EEEE
turns TTT TTTTTT TT TTT
coil CC CC CCC
. 70 . 80 . 90 . 100 . 110 . 120
GVSLTKLAPQSHFGNIIVFDDPITLSHSLSSKQVGRAQGFYIYDTKNTYTSWLSFTFVLN
helix HHHHHH HH
sheet E EEEEEE EEEEEEEEEE EEEEE
turns T TTT TT TTT TTTTTTTT
coil CC C C CCCCC CCCC
. 130 . 140 . 150 . 160 . 170 . 180
STHHQGTITFAGADPIVAKTRDISVTGGTGDFFMHRGIATITTDAFEGEAYFRLGVYIKF
helix HHH HHHHHHHHH
sheet EEEE EEEEEEEE EEE EEE EEE
turns TT TTT
coil CCCC CCCC CCCCCC CCCCC CCC
FECW
helix H
sheet E
turns TT
coil
Residue totals: H: 38 E: 66 T: 38 C: 42
percent: H: 22.6 E: 39.3 T: 22.6 C: 25.0
DRR206 is strongly induced by Fusarium oxysporum in seedlings of
near isogenic pea lines Vantage (R) and M410 (S).
Hadwiger et al. (1992) Phys. Mol. Plant Pathol. 40:259-269.
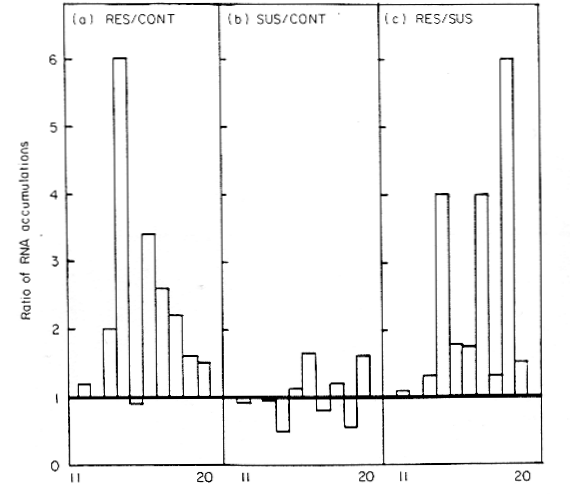
DRR206 genes are most strongly-expressed
against incompatible races of Pseudomonas syringae pv. pisi.
Daniels et al., (1987) Plant Mol. Biol.
8:309-316.
| Pea cultivar |
P. syringae race |
Infection phenotype |
Time period |
| 10h |
20h |
30h |
| Mars |
R-1 |
S |
1.52 |
1.46 |
0.92 |
| R-2 |
R |
1.86 |
4.95 |
1.98 |
| R-3 |
S |
2.26 |
1.13 |
1.61 |
| Abador |
R-1 |
R |
2.10 |
5.11 |
2.01 |
| R-2 |
R |
1.98 |
3.41 |
2.05 |
| R-3 |
S |
1.33 |
3.43 |
2.12 |
| Ceras |
R-1 |
R |
4.23 |
3.37 |
2.15 |
| R-2 |
S |
1.72 |
2.33 |
1.51 |
| R-3 |
S |
1.56 |
2.09 |
0.75 |
| Mini |
R-1 |
R |
2.85 |
4.77 |
3.60 |
| R-2 |
S |
1.54 |
2.45 |
2.71 |
| R-3 |
S |
2.31 |
2.89 |
3.23 |
| Spring |
R-1 |
S |
1.12 |
1.91 |
1.19 |
| R-2 |
R |
0.81 |
4.64 |
1.47 |
| R-3 |
R |
0.38 |
2.02 |
1.12 |
Signal from Northern blots expressed as a fraction of water
control.
DRR206 is also induced by lipoprotein-peptidoglycan preparations,
or killed cells, from P. syringae in a non-race-specific fashion.
Daniels et al., (1988) Phytopathology 78: 1451-1453. |



