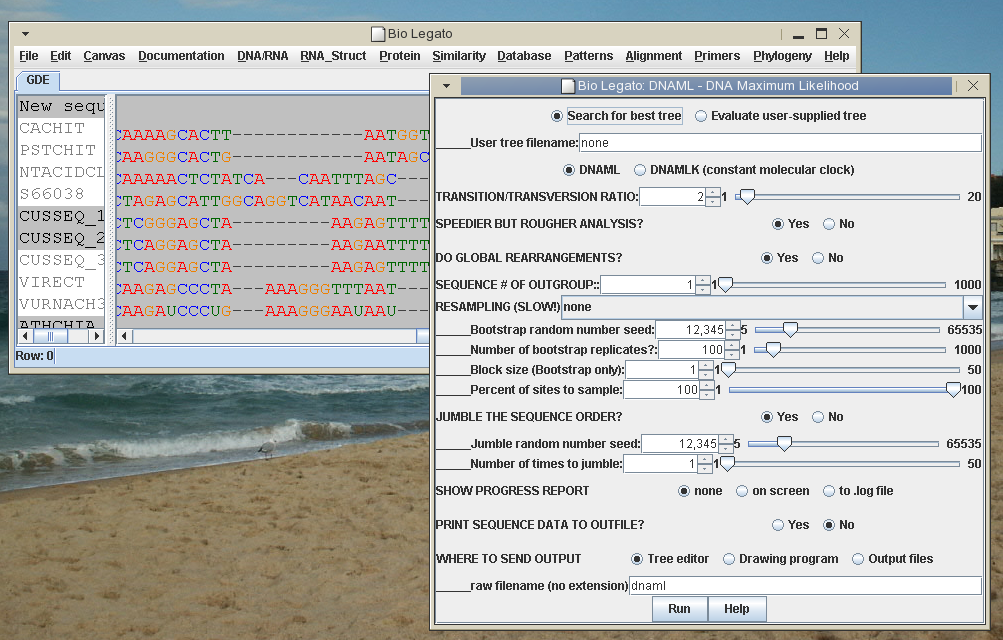
Figure 1 - bioLegato main window containing a
multiple DNA sequence alignment
and a
popup menu for launching the DNAML program.
bioLegato
is a programmable graphic interface capable of handling a wide range of
data types. bioLegato is:
bioLegato takes its initial inspiration from Steven Smith's GDE
interface.
Each
bioLegato window consists of two panes, the
menus (methods) and the canvas (data), as demonstrated in Figure 1. In
bioLegato, data is selected in the canvas pane and then a
program is chosen from the menu pane. A popup menu lets the user set
program parameters and then launch the program. When the program has
completed, the output pops up in a new window.
 |
|
Figure 1 - bioLegato main window containing a
multiple DNA sequence alignment
and a
popup menu for launching the DNAML program. |
Any number of canvases may be plugged into the canvas pane, each specialized for displaying, manipulating, and selecting data items of a specific type. Data types might include any data retrieved by Seahawk: microarray data, DNA or protein sequence data, phylogenetic trees, or protein structures. For each instance of a bioLegato window, menus appropriate for a given canvas will appear in the menu pane. For example, a bioLegato protein object would have protein sequences on a protein canvas and menus for launching protein-sequence analysis tasks would appear in the menu pane. The actual tasks that appear in the menus could be either BioMoby web services or locally-installed standalone programs. Output could be displayed using viewers for bitmap graphics (e.g. JPEG), HTML, sequence alignments, or protein structure viewers. If the object was another genome, a new instance of Bluejay might be called. Most importantly, if the output is of a type that can be worked with by bioLegato, the output could also pop up in a new bioLegato instance. In that way, bioLegato allows the user to interactively create an ad hoc data pipeline.
All
tasks run by bioLegato are called as external programs rather than
being hard-coded into bioLegato itself. The menus to call these
programs are read from a menu-spec file when bioLegato is launched.
In principle, any locally-installed program or remote web service can
easily be added to bioLegato by adding an entry to the menu-spec
file.
Our first goal, to make bioLegato a useful replacement for GDE in the BIRCH system, is now essentially complete. In the long term, bioLegato will be an ideal interface for almost any set of programs for working with a particular type of data. In the longer term, as more canvases are developed, the range of data types that can be handled by bioLegato will broaden. From a developer's point of view, development of new applications programs could become as simple as developing a new canvasand menus for a specific data type. At the same time, bioLegato will help to integrate many of the existing components of the Bioinformatics Platform into a more uniffied system.