
CHEM 4620
Biochemistry of Nucleic Acids
CHEM 4620 Biochemistry of Nucleic Acids
(3)
The structure of nucleic acids; synthesis and sequence determination; interaction with drugs and protein.
Not to be held
with the former 002.453. Prerequisite: CHEM 2370 (MBIO 2370) (or the former 002.235 (060.235))
Please read this important notice from the
Faculty of Science:
Registration advisory
2007-2008
Course Outline - 2009 (CURRENTLY UNDER REVISION)
Instructor:
- Dr. Sean A. McKenna
- Room 380 Parker Chemistry Building
- Office Hours: or by appointment.
- Telephone: 474-
- E-mail: mckenna@cc.UManitoba.CA
Lectures:
- Mon, Wed, & Fri 9:30-10:20
AM
- 539 Parker
- No laboratory.
Textbook:
- None. Short journal articles are required reading, in addition to the notes & handouts.
- Series A, etc. refer to collections of handouts containing structures, mechanisms, 3D diagrams, etc, relevant to the various topics.
Sheets:
- Sheets with structures and tables handed out in class may be required for
several lectures. You must bring these sheets to class; lectures cannot
be understood without them.
Review Sheets:
-
Review sheets with questions will be handed out. Some questions on the
midterm and final exam will be taken from these sheets, so work through
them. You should also work through old exams that may be available at:Online Examinations.
Grading:
- Mid-term Test 1 TBA (in class) 50 min during lecture slot.
-------- 20%
- Mid-term Test 2 TBA (in class) 50 min during
lecture slot.
-------- 20%
- Final Exam 3 Hours. Scheduled by
Student Records ---------------- 60%
Academic Dishonesty: Please visit
the Faculty of Science web site
Cheating, Plagiarism etc.
Important information in the 'General Calendar':
-
Students are reminded
that they must remain available until all examination and test obligations
are fulfilled.
- Deaths in
the immediate family and illness are the only valid reasons for missing
tests and exams. A medical certificate must be provided as proof of
illness. If a midterm is missed, the final exam will be valued at 80%
towards the student's final grade.
Course Description
Abbreviations: BP, base pair. CAP, catabolite gene activator protein. CD, circular dichroism. DH, double helix. DHical, double helical. HBing, hydrogen bonding. HBs, hydrogen bond(s). LH & RH, left & right hand. NOE, nuclear Overhauser effects. PUR, purine. PYR, pyrimidine. RNase, ribonuclease. SS, single strand. W-C, Watson-Crick. XRD, x-ray diffraction.
Series A 5.5 Lectures
- Properties of PUR & PYR Bases:
- Review
: Base, nucleosides, oligomers & polymers.
- Introduction to XRD: A-, B-, Z-DNA forms are apparent from fibre diffraction patterns.
- Bond angles & lengths: from single-crystal XRD data.
- Planarity of the bases: puckering of 5,6-saturated PYRs - important in tRNA, UV-damaged DNA.
- &Pi-bond structures & Tautomerism.
- Acid-Base Properties
: Ionization of Pur & Pyr bases in the pH range 0-12. Ionization of the phosphate & the sugar 2'-hydroxyl of ribonucleosides - Ribozymes.
- Role of the 2'-OH in RNA Hydrolysis:Introduction to the RNA World.
- In Basic Solution: Does not occur in DNA.
- RNase A: at (Pyr)pN sites - His residues involved in catalysis.
- Leadzymes: Pb2+-catalyzed hydrolysis of tRNA backbone at D17pG18. Insight into role of M2+ ions.
- Ribozymes & RNA Splicing: Mechanism of removal of Group II introns & lariat formation.
- Ribosomes are Ribozymes: Peptidyl transferase - role of an adenine with unusual pKa.
- Chemical Reactivity of PURs & PYRs:
- Electrophilicity & Nucleophilicity of C & N sites of Pur & Pyr bases.
- C5 atom: A) Mechanism of Thymidylate Synthase: Folic acid, hydride transfer, tritium labeling. Antitumor agents, 5-fluorouracil, methotrexate.B) Methylation of Cyt: S-adenosylmethionine, SAM.
- C4 atom: Cytidine Deamination in Acid & Basic Solution: Source of transition mutations.
- Enzymatic Deamination of Cyt, Ade & Gua: Zn2+-containing enzymes.
Series B 4 Lectures
-
Hydrogen-Bonding by PURs & PYRs
- BPs: Watson-Crick, Reverse WC, Hoogsteen, Reverse Hoogsteen.
- Transitions & transversion mutations. Base tautomerism, ionization, wobble.
- Mutations induced by carcinogens: alkylating agents, SAM, etc.
- UV Absorption & Base Stacking: Relevant to CHEM 4700 lab.
- Beer-Lambert Law (BLL): absorbance & extinction coefficients (&epsilon), spectra of U, C, T, A, G: effect of pH change. Analytical uses.
- Deviations from BLL due to BSing. Hypochromic effects due to diminished &epsilon of bases.
- BSing forces: dipolar & London dispersion forces, role of solvent.
- UV absorption & BSing in oligomers: hypochromic affects of chain extension; hypochromicity; hyperchromic effects of temperature increases. Melting curves & melting temperature (Tm) provide insight into BSing: nature of base (Pur or Pyr), solution pH, base methylation, chain length, solvent, disruption by 5,6-reduction of Pyr bases.
- Melting curves & Tm of DHical DNA: G/C content. DH stabilized by Mg2+, destabilized by Cu2+.
Series C 2.5 Lectures
- Klyne-Prelog proposal for torsion angles in A-B-C-D fragments; definition of syn, anti, gauche+, trans, & gauche- conformers.
- Defining the torsion
angles of a nucleotide unit of RNA & DNA: A) 6 backbone bonds, &alpha, &beta, &gamma, &delta, &epsilon, &zeta (zeta); B) 5 furanose ring bonds, &nu 0 -> &nu 4 (define sugar pucker); C) syn & anti ranges of the N-glycosyl bond, the &chi (chi) angle; syn bases are present in LHed Z-DNA.
- The endo/exo nomenclature The 2'-endo & 3'-endo puckers define the B- & A-families of DNA & RNA helices, respectively. Why the 2'-endo pucker leads to a more extended (B-type) DH.
- Proton NMR (PMR) for solution studies of stereochemistry: PMR spectra for small oligomers. Data derived from spectra: peak intensities, chemical shifts, coupling constants (J), relaxation times. Classes of J: geminal, vicinal, long-range.
- The Karplus Equation: structural analysis of nucleic acids, proteins, carbohydrates.
- Sugar ring pucker: Experimental J data reveal flexibility, i.e, interconversion between 2'-endo & 3'-endo puckers. Data for stacked oligomers suggest less flexibility.
- The power of the XRD-NMR combination of techniques for structure determination.
Series D 4 Lectures
- History: The 2 W-C 1954 papers are required reading. Short biographies of Watson, Crick, Wilkins, Franklin. LHed & RHed helices; 3D diagrams are examined.
- RHed A- & B-DNA: A) Similarities: Antiparallel chains; +ve twists; anti bases; propeller twists; major & minor grooves; Pur & Pyr ring atoms exposed in the grooves. B) Differences: sugar pucker is 3'-endo in A, 2'-endo in B; the relative orientation of adjacent base-pairs (BPs) as defined by the extent of their 'twist", 'roll', & 'slide', and BP inclination. Dramatic differences in width & depth of the minor & major grooves of A- & B-DNA; accessibility of grooves to proteins, etc.
- XRD reveals impact of hydration on DNA fibres.
- LHed Z-DNA:A) Early CD & P31-NMR studies reveal unusual behaviour of DNA at high salt concentration. B) 1979 Rich & Dickerson single-crystal XRD work on C/G oligomers reveal LHed DNA with a 'zig-zag' orientation of P atoms. C) Details of Z-structure: Syn G bases but anti C bases; -ve twist of BPs; dramatic change in groove dimensions. D) XRD data explain the unusual behaviour in (A). E) NOE data provide support for presence of Z-DNA in solution.
- Summary of structure determination by NMR techniques.
- Does Z-DNA have a biological role? Alexander Rich's Proceedings of the National Academy of Science 102, 12759 (2005), "Biological function of the vaccinia virus Z-DNA-binding protein E3L: Gene transactivation & antiapoptotic activity in HeLA cells.
Series E Chiroptical Methods: CD & ORD (Optical Rotatory Dispersion) 3.5 Lectures
Provides background for understanding CHEM 4700 experiments on nucleic acids & proteins.
- Properties of light: Amplitude & wavelength (frequency).
- 'Space" & 'Time' dependence of A) plane (linearly) polarized light (PPL) & B) circular polarized light (CPL). Right & Left CPL. Superposition of RCPL & LCPL yields PPL.
- Sources of molecular chirality: chiral atoms, helices, supercoiling. Chirality in nature.
- Optical activity of a solution: chiral molecules interact differently with LCPL & RCPL, ie, in the solution, their molecular extinction coefficients differ (CD) & velocities (refractive indices) differ.
- Vector analysis of the effect of "optically active solutions" on incident PPL: A) PPL is rotated clockwise (dextrorotatory solution) or counterclockwise (levorotatory);B) PPL is converted to "right elliptically polarized light" or "left elliptically polarized light".
- CD spectroscopy: A) Ellipticity of optically active solutions.B) Positive & negative cotton effects. C) CD spectra (UV range) of nucleosides, oligonucleotides, DNA; effect of temperature, solvent & pH. D) CD & protein structure.
- Required reading: "Mirror-Image DNA". CD spectra are provided for d(CGCGCG) containing either the normal D-sugars or L-sugars. Unusual structures such as RHed Z-DNA are noted. This illustrates - what might seem, at first sight, a paradox - that, although the bases are the light absorbers, the non-absorbing chiral sugars invert the CD spectra.
Series F UV Damage & Thymine Dimer Formation
1.5 Lectures
- Thymine Dimers: Cell death, mutations, skin cancer. Repair by photoreactivation & excision. UV absorption studies of Tm lowered; 'bubble' formation.
- XRD study by Cadet, Hruska, et al of the d(TpT) photodimer, the 1st such study of a thymine dimer with the sugars attached. Dua reveal the non-planarity of the damaged Thy bases & dramatic changes in the &chi (chi) angle, the sugar pucker, and backbone bonds (&alpha, &beta, etc). Relevance to recognition by repair enzymes. PMR data of Hruska et al reveal distortions in solution.
- XRD structures of oligomers containing a thymine dimer.
Series G tRNA Structure
6 Lectures
- Primary (1o) structure: SSed; Major (A, G, C, U) & modified units. Modifications fine tune roles of tRNA - specificity of amino acid acylation, codon reading, translation activities.
- Cloverleaf (2o) structure: Stems & loops. Mismatched BPs. Conserved & semi-conserved units.
- Overall 3-Dimensional structure Based on XRD data for tRNA(phe).
- L-shaped: arrangement of stems & loops in the 2 limbs of the L.
- 2o features of stems: A-type DHs.
- 3o Structural forces:
- HBs between distant stems & loops. Unusual BPs. Base triples involving the D-stem.
- Base-to-2'-OH HBs: not possible in DNA.
- 3o BSing: Intercalation involving distant bases.
- Role of bound water: bridging between 2'-OH & O2 (Pyr) or N3(Pur).
- Role of Mg2+: stabilizing sharp bend by bridging phosphates near U8, etc.
- Evidence that tRNA structure is similar in solution & the crystal state
- Pb2+ catalyzed hydrolysis of tRNA is consistent crystal folding pattern.
- UV-induced cross-linking of bases in different tRNA segments.
- Chemical probes: P-32 radioactive labeling, specific base modifications, chemical strand scission, separation of oligonucleotides by PAGE.
- Miscellaneous
- tRNA binding to cognate aminoacyl tRNA synthetase (XRD Steitz group).
- Macromolecular mimicry: Structure of the ternary complex of EF-Tu.
Series H Introduction to Protein-Nucleic Acid Binding
1 Lecture
- Importance of Protein Interactions
- Specificity of binding
- To SSs or DHs. Base sequence.
- Review of protein structure: amino acids (non-polar, etc), &alpha-helix, &beta-sheet.
- Potential interactions: Salt bridges, HBing, BSing, hydrophobic.
- Hypothetical interactions with bases of SSs (all donor & acceptor sites on the base are accessible for HBing) or of DHs (W-C sites are protected & the base planes are not available for BSing). Interactions within the grooves.
- BP recognition codes for interactions in grooves. Restriction enzymes codes.
Series I The RNase T1-2'-GMP Complex
1 Lecture
XRD & solution data provide insight into protein binding to SS RNA & i
nto reaction mechanisms.
-
General Information: T1 hydrolyzes specifically at G units. 2'-GMP is a competitive inhibitor.
- Structure of T1: 1o (Sequence known; 2 S-S bridges). 2o (4.5 turns of &alpha-helix. 7&beta-strands. Loops). 3o (Globular. Distribution of basic & acidic R groups. 2'-GMP is directed to binding site on T1 surface). 2'-GMP structure is revealed by the XRD data
- 2'-GMP binding to T1
- 6 HBs to the G base: 2 with an R-group, 4 with backbone amides.
- Partial intercalation of G base between Tyr 42 & Tyr 45. O6 & N1 of G protected from solvent.
- Proposed Mechanism of T1 Catalysis: Based on XRD structure, solution data, genetics. Involves transesterification, followed by hydrolysis of the 2',3'-cyclic phosphate. Glu & His catalytic residues. Comparison with the RNase A reaction in Series A.
Series J CAP Binding to DNA
2.5 Lectures
Illustrates sequence-specific protein binding to DHical DNA & DNA bending.
-
1o
Structure of CAP: Dimer with 2 identical subunits.
- Structure of the cAMP-CAP Complex
- CAP Subunit Structure: 2o: 6 &alpha-helices (A-F) & 12 extended &beta-strands (1-12). 3o: Folding forms N- & C-domains separated by a 'hinge'. cAMP binds to an antiparallel &beta-barrel (strands &beta 1 -> &beta 8).
- 4o Structure of CAP Dimer: Subunits held mainly by hydrophobic forces between their N-domains; &alphaF helices of the C-domains project out from dimer surface.
- cAMP Binding to CAP: 8 HBs & 1 salt bridge involving base, sugar & phosphate groups of cAMP, & acceptors/donors on the R & backbone groups of CAP.
- Methods Used to Identify CAP Binding Site on the Promoter of the lac Operon
- DNase protection; phosphate ethylation; base methylation; UV-induced crosslinking; DNA mutants.
- A near-palindromic site of ~26 BPs.
- Discussion of XRD Data for the Ternary cAMP-CAP-DNA Complex
- Kinks formed by large "roll angles" between BPs 5 & 6 on each side of the "2-fold" axis of symmetry. ~90o bend in the DNA DH.
- CAP binds in major groove of B-DNA. Discussion of the specific CAP-DNA interactions.
- Other major-groove binding proteins.
Series L Netropsin Binding to DNA
1.5 Lectures
Illustrates binding of a drug in the minor groove of B-DNA.
-
Structure of NET: two N-methyl pyrrole & 3 trans planar amide groups, +vely charged end groups.
- NET-DNA Binding in Aqueous Solution
- Specific for A/T regions of B-DNA. No affinity for A-, Z- or SS DNA.
- Evidence for groove binding: W-C BPs remain intact. No unwinding or lengthening of DH.
- Evidence for minor groove binding: base methylation; intermolecular NOE measurements.
- XRD Data for Unbound NET: Flat, crescent-shaped. N-H groups project out from concave edge.
- XRD Data for the DNA-NET Complex
- NET favours A/T rich regions. Consistent with solution studies.
- Minor changes in NET structure: NET was designed for DNA binding!
- DNA remains in B-form: Minor groove expands slightly at NET site.
- Spine of Hydration in minor groove of B-DNA. Propeller twists of BPs; HBing with base edges.
- H2O molecules from minor groove at NET site. NET mimics the spine of hydration.
- Reason for A/T specificity: steric hindrance by 2-NH2 of Gua.
- Thermodynamic data: +ve entropy change for NET binding due to release of many H2O molecules.
Series M In Vitro DNA Synthesis
1 Lecture
- Because of expansion of earlier topics, this topic has not been completed in recent years. In 2005 I managed to cover only a comparison of the chemistry of in vitro & in vivo synthesis, & the importance of blocking & activation of starting materials prior to in vitro polymerization.
Miscellaneous Information
- Lecture distribution: 34 lectures & 2 in-class tests.
- Number of Students who completed the course (GPA):
2005 26 (3.44). 2004 (36, 3.03). 2003 21 (3.41). 2002 25 (3.40). 2001 22 (3.43). 2000 24 (3.23)
- 5-Year average enrolment: 26
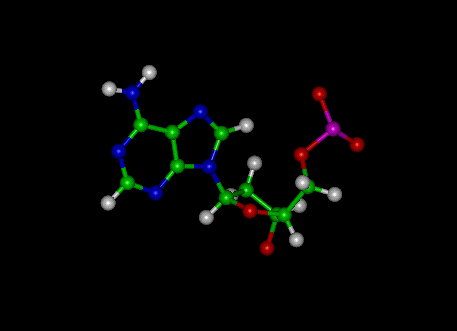
Adenine
|
Thymine
|
 |
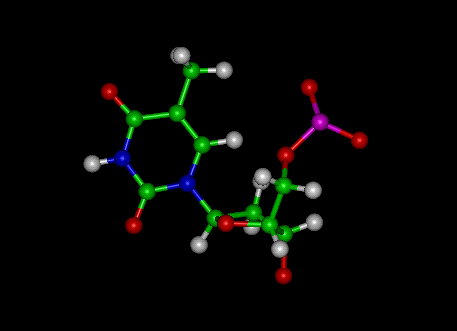 |
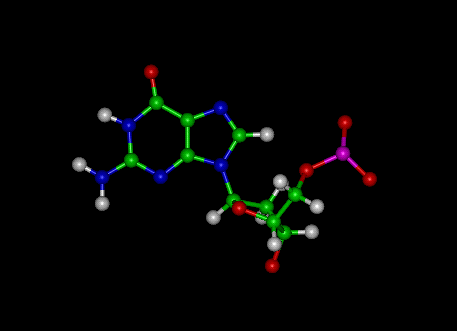
Guanine
|
Cytosine
|
 |
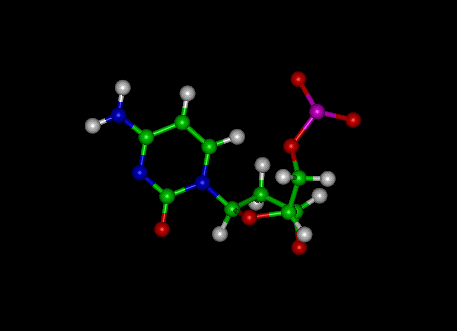 |
Uracil
|
DNA!
|
 |
 |
Useful Web Sites for CHEM 4620:
Nucleic Acid Database
Return to the Chemistry Department Course Descriptions
Return to Dr. McKenna's Home Page
http://home.cc.umanitoba.ca/~joneil/CHEM4620.Course.outline.htm
Maintained by J. O'Neil

Free JavaScripts provided
by The JavaScript Source









