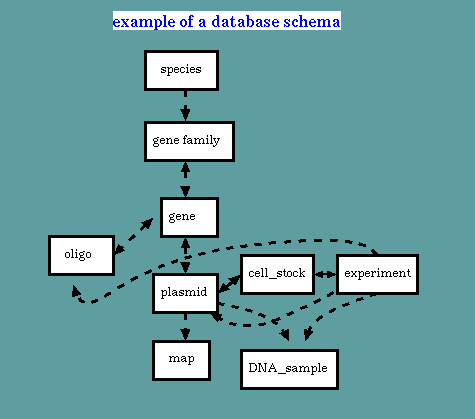
A database can be simply a repository of data. However, by encoding the relationships between different types of information, it becomes a knowledge base. In the figure above, information about different types of objects, such as species, plasmids, or DNA samples, is stored in discrete entries, known in ACEDB as 'classes'. Links between classes, called 'tags', show how they are related. For example, a species may have many gene families, and a gene gamily may be represented by many copies of a gene. An experiment may point to many types of objects.
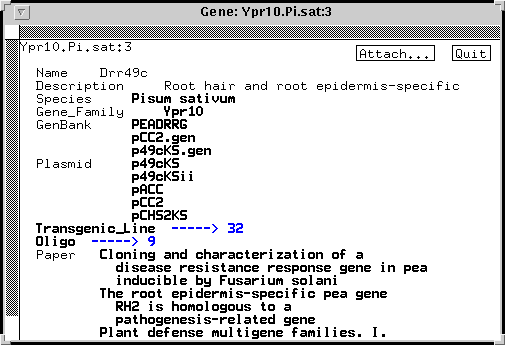
In ACEDB, these relationships can be implemented by defining classes tailored to the needs of your lab. For example, one copy of the PR10 gene from pea might be represented thus:

This object is a discrete entity in and of itself. At the same time, all boldface tags point to objects in other classes. So, from a gene, you could click on a tag and go to the species from which it is derived, or to the gene family to which it belongs, or to GenBank entries, to plasmid constructs containing the gene, to transgenic plant lines contianing the gene, to oligonucleotides specific for that gene (in this case, there are so many oligos that you have to click on the '---->9' to display the list), or to papers in which the gene was studied.
In another example, we see that the specificity of an oligonucleotide can be represented by pointing to genes:
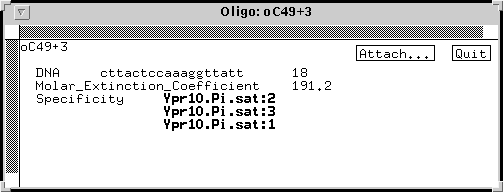
Oligo oC49+3 was designed from a region that is conserved among 3 copies of the pea PR10 gene. Thus, it lists 3 copies of the gene, each of which could be amplified using this oligo as a primer.
The main window serves as a starting point for browsing the database:
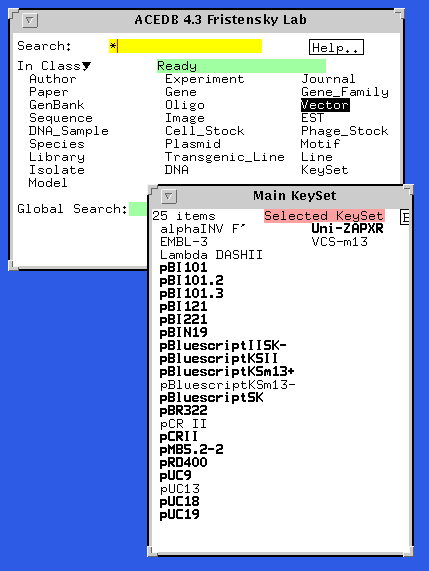
The main window displays all the major classes of objects encoded in the database. Any class can be a starting point. In this example, the user has clicked on 'vector' to display a list of vectors used in the lab. Clicking on any of these vectors will display the entry for that vector.
NEXT: