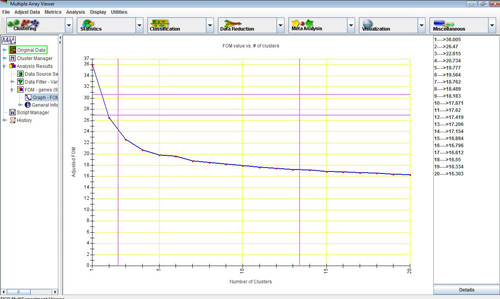
FOM vs. No. of Clusters graph for KMC algorithm. (1-20 clusters, 20 iterations)
(Yeung et al. 2001)
The Figure of Merit is, in concept, a measure of fit of the expression patterns for the clusters produced by a particular algorithm. MeV's FOM implementation provides FOM results for running the KMC and CAST clustering algorithms. Each algorithm is initialized by selecting either the K-Means/K-Medians tab or the CAST tab.
Currently, FOM is available for the CAST, K-means and K-medians algorithms. A figure of merit is an estimate of the predictive power of a clustering algorithm. It is computed by removing each sample in turn from the data set, clustering genes based on the remaining data, and calculating the fit of the withheld sample to the clustering pattern obtained from the other samples. The lower the adjusted FOM value is, the higher the predictive power of the algorithm. The "Maximum number of clusters" input field under the K-Means / K-Medians tab in the initialization box is used to determine how many times FOM values should be calculated for the k-means/k-medians algorithm.
Each time, the number of clusters computed is increased by one, starting with one cluster in the first iteration. The "Interval" input field under the CAST tab allows the user to specify the increase in threshold affinity in successive iterations of CAST. If the "Take Average" box is checked, in case there is more than one clustering outcome for a given number of clusters, the average FOM for that number of clusters will be used to draw the FOM-vs.-number of clusters curve. If the "Take Average" box is unchecked, each FOM value will be represented in case of such a tie, and curves will be drawn through each value.
In the figure below, the value of the adjusted FOM for a K-means run decreases steeply until the number of clusters reaches 4, after which it levels out. This suggests that, for this data set, K-means performs optimally for 4 clusters and that any additional clusters produced will not add to the predictive value of the algorithm. FOM is useful in determining the best input parameters for a clustering algorithm.
This field specifies a number of FOM iterations to run during the analysis. When the K-Means mode is selected and the number of FOM iterations is greater than one, the mean FOM values are reported with the standard deviation on the output graph. This is useful in the case of K-means where the initialization step involves an initial random partitioning. Each K-means run is potentially unique although each should be similar. When using this option the result can be based on several runs. A right click in the graph will provide the option to show or hide the individual lines representing FOM iterations.
Threshold Interval
For FOM an interval is used to perform a series of CAST runs in which the Affinity Threshold is incremented from 0.0 by the interval indicated. The default of 0.1 is often a good value since it provides 11 CAST results from 0.0 to 1.0 incremented by 0.1.
The threshold parameter is a value ranging from 0.0 to 1.0 which is used as a cluster affinity threshold. Each expression element will have an affinity for the current cluster being created based on it's relationship to the elements currently in the cluster. If that affinity is greater than the supplied threshold the gene is permitted to be a member of the cluster.
Means/Medians Option
The Means or Medians option indicates whether each cluster's centroid vector should be calculated as a mean or as a median of the member expression patterns.
Maximum Number of Clusters
This positive integer value indicates the maximum number of clusters to be created. For instance, if the entered value is 10 then KMC is run 10 times to produce 1,2,3..,10 clusters. An FOM value is returned for each run.
Maximum Number of Iterations
This positive integer value is the maximum number of times that all the elements in the data set will be tested for cluster fit within a single KMC run. On each iteration each element is associated with the cluster with the closest mean (or median). Note that a KMC run will terminate when either no elements require migration (reassignment) to new clusters or when the maximum number of iterations has been reached.