VISA is a tool for finding amino acid motifs that are conserved within a set of protein sequences. VISA identifies amino acid patterns that are common to many members of a set of amino acid sequences, and displays the distribution of common patterns along the sequences in a series of histograms. Individual peaks of these histograms can be assigned different colors. Common sequence patterns inherit and display the color of the peak in which they occur, leading to analogous segments in the other sequences being marked in matching colors. These peaks usually correspond to the conserved sequence motifs that are characteristic of the studied proteins. The resulting color graphic overview of sequence similarities can help to understand the architecture of the protein family and can be helpful while designing experiments to probe function.
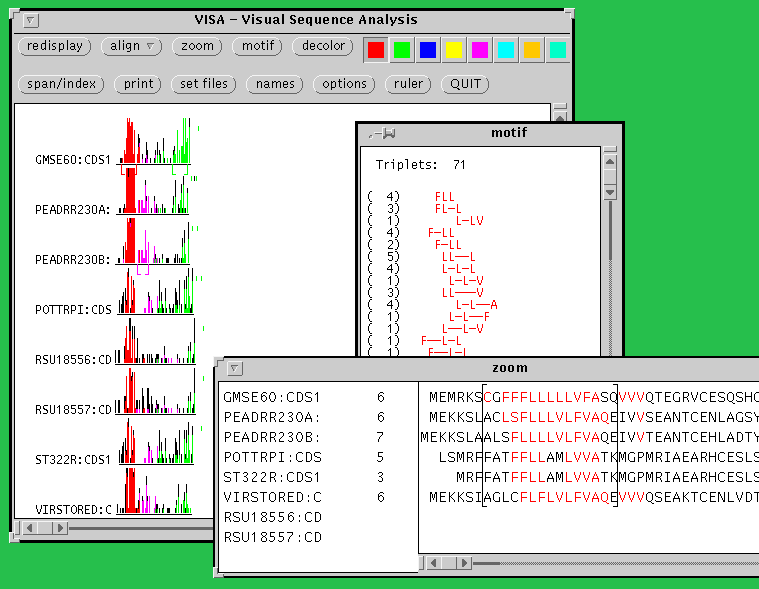 Documentation for VISA can be found in
Documentation for VISA can be found in
VISA can be launched from the command line by typing 'visa &', from the OpenWindows Workspace menu under 'Molecular Biology', or from GDE in the Patterns menu.