Coloring Cells for Alignment
Manual alignment can be made easier by appropriate choice of a method to color
cells. You can set the method to color cells using the Matrix>Color cells
submenu. Three useful methods are:
- Character State — This colors cells according to
nucleotide. This is the default coloring, but in case it has been turned off,
you can turn it back on by going to Matrix>Color Cells>Character State.
- Aligning Colors — This colors pyrimidines by similar
colors, and likewise purines
- Highlight Apparently Slightly Misaligned — This highlights
subsequences that appear better shifted one or two sites left or right:
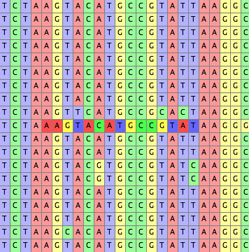
It bases its calculations on the dominant nucleotides in the columns, and
hence is useful only in a matrix that is mostly well aligned. Shifts more
distant than one or two sites are not considered. Do not consider this a definitive
indication that the cells should move; its purpose is only to help you find
possible issues, but the decision as to whether to shift or not is still up
to you. The highlight does not specify which direction the subsequences might
be best shifted; to learn this, hold the cursor over the sites in question.
Selecting Hint
To select from one cell to the end of the sequence, select the cell and then
hit Shift-Alt-Arrow key (or, on OS X, Shift-Option-Arrow key), where the arrow
key is either the right or left arrow key.
Preparing the Matrix
Several options under Matrix>Alter/Transform may be useful to modify the
matrix in preparation for alignment. Some of these options are under "Other
Choices...".
- Collapse Gaps to Left
- Collapse Gaps to Right
- Trim Terminal Gaps Character
- Gaps to Missing
- Missing to Gaps
- Terminal Gaps to ?
- Terminal ? to Gaps
- Reverse Complement
- Reverse
- Nucleotide Complement
