GDE Menu Overview
Menus provide a way of organizing available programs to
make them easier to find, and present the choices available.
The greatest strength of GDE is that programs can be added to the
menus by inserting a few lines of code into the .GDEmenus file, which is read each time
the program starts. As new programs are added, or existing menus
changed, the changes are immediately available to all users
when GDE starts up. Thus, the menus shown below may not
include all programs available.
File menu
The file menu is used for importing and exporting sequence
data in various formats.
Open - GenBank, gde flat file and gde format files.
Import Foreign Format - most other file types
Open_XYLEM - XYLEM datasets (see splitdb)

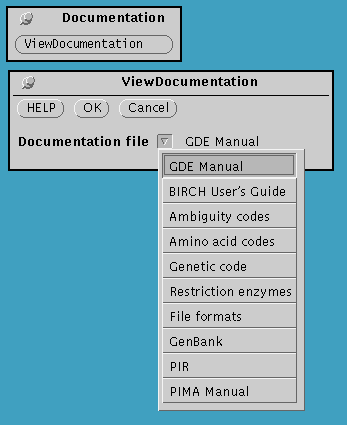
Documentation menu
Click on ViewDocumentation to get a pull-down
menu listing useful documentation files.
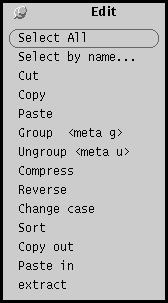
Edit menu
The Edit menu lists sequence editing tasks. Copy out copies the
selected sequences to a file called $HOME/.GDEclipboard. Paste
in pastes the contents of that file into the current GDE
window. Therefore, Copy out/Paste in can be used to move
sequences from one GDE window to another.
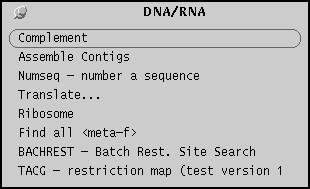
DNA/RNA menu
The DNA/RNA menu lists programs for common sequence tasks,
such as printing, translation, complementation, and
restriction site analysis.
RNA structure menu
The RNA structure menu lists programs for RNA
structure prediction.

Protein menu
The protein menu lists programs for structure
prediction, amino acid characterization and
reverse translation.
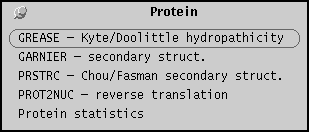
Similarity menu
The similarity menu lists programs for pairwise
comparisons of DNA or protein sequences, using
either global alignment or dot-matrix methods.
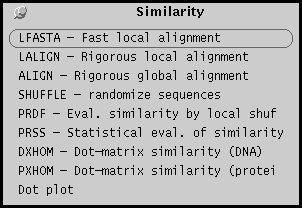
Database menu
The Database menu lists programs for searching databases using
either keyword or sequence information, and for constructing
database subset tailored to the user's specifications.
Patterns menu
The Patterns menu lists programs that facilitate
the identification of motifs conserved among
a group of sequences.

Alignment menu
The Alignment menu lists programs for construction of
multiple sequence alignments, as well as programs
for display of multiple alignments.

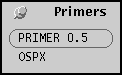
Primers menu
The primers menu lists programs for primer design.
Phylogeny menu
Most programs in the phylogeny menu expect that three of
more sequences, aligned by one of the programs in the alignment
menu, have been selected. In most programs, output will consist
of a report and a treefile. Most menus allow both files to
be saved directly to disk (recommended for long running
jobs) or to display directly using TREETOOL, DRAWTREE or
DRAWGRAM. These three programs can also read in treefiles
from disk. Finally, the Phylip help menu item displays
individual Phylip documentation files in a textedit window.
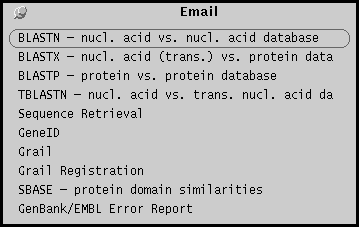
Email menu
The email menu lists programs that can be run by
email requests to other sites. The selected sequence(s) and
parameters are inserted into a mail message which is sent
to the remote site. The results return to the user via email.
To use GenBank/EMBL Error Report, type in an Accession
number. The GenBank entry will be retrieved to one textedit
window, and the Error Report form to another window. Using
the entry for reference, the error report can be filled
in. After saving and quitting the Error report file, the
file will automatically by emailed to GenBank or EMBL.
An acknowledgement is returned automatically by email within minutes,
and the decision of the database annotators will usually
be forwarded within a few days.