The default BIRCH distribution runs BLAST by sending jobs to the BLAST server at NCBI. However, part of the design of BIRCH is to make it easy to add local programs to a BIRCH system. At the COE, BLAST searches are accelerated using the Paracel Blast server. PcBLAST searches can be run from the Visual Genomics web site, or from GDE at the COE BIRCH site.
In this tutorial, we will demonstrate BLAST searches using some of the defensin sequences uploaded in a previous demo. Alternatively, you can directly download the file defensin.CDS.gde from this page.
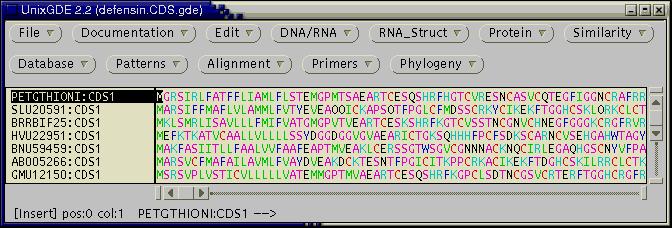
Open a terminal window and cd into the directory containing defensin.CDS.gde. Now, open this file in GDE
gde defensin.CDS.gde

To demonstrate pcBLAST, select any sequence, and choose Database --> Paracel BLASTP. The following menu will appear.
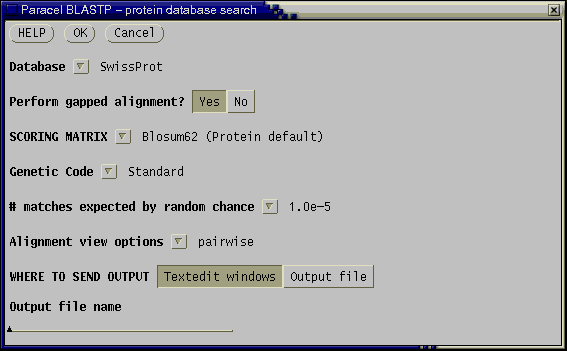
Set Database to SwissProt, and the number of matches expected by chance to 1.0e-5 (ie. e = 1.0 x 10-5). Click on OK and the search will begin. In the terminal window in which gde was launched, progress messages will be written. When the output is complete, it will appear in a text editor. In this example, the SwissProt database was searched with PETGTHIONI protein sequence. The results can be found in THG_PETIN.pBLASTP.