TUTORIAL: FINDING AND RETRIEVING COMPLETE EUKARYOTIC GENOMES |
Nov. 22, 2023 |
TUTORIAL: FINDING AND RETRIEVING COMPLETE EUKARYOTIC GENOMES |
Nov. 22, 2023 |
mkdir getgenomeFor genome downloads, the user may choose one or more files with sequence and annotation in various formats. For this reason, files are distributed in zip archives which include subdirectories and metadata files. Since this tutorial only requires the FASTA sequence file, create a temporary directory for the download that will make it simpler to delete the remaining files later.
cd getgenome
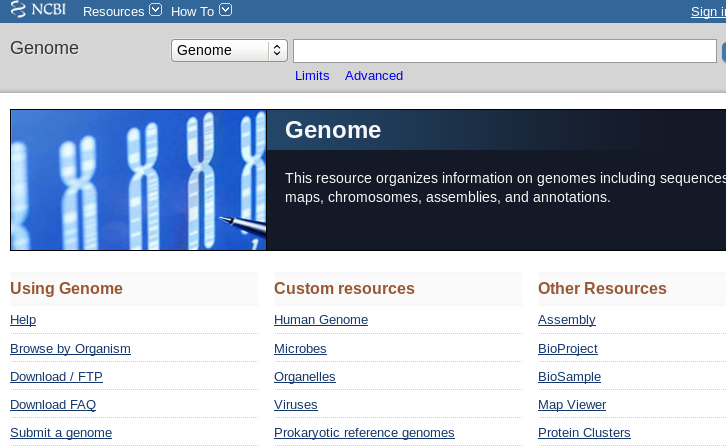
| The best starting point is the Genome page at
NCBI [https://www.ncbi.nlm.nih.gov/genome]. Click on Browse by Organism and type the genus name "Saccharomyces" into the Search by organism box.
|
 |
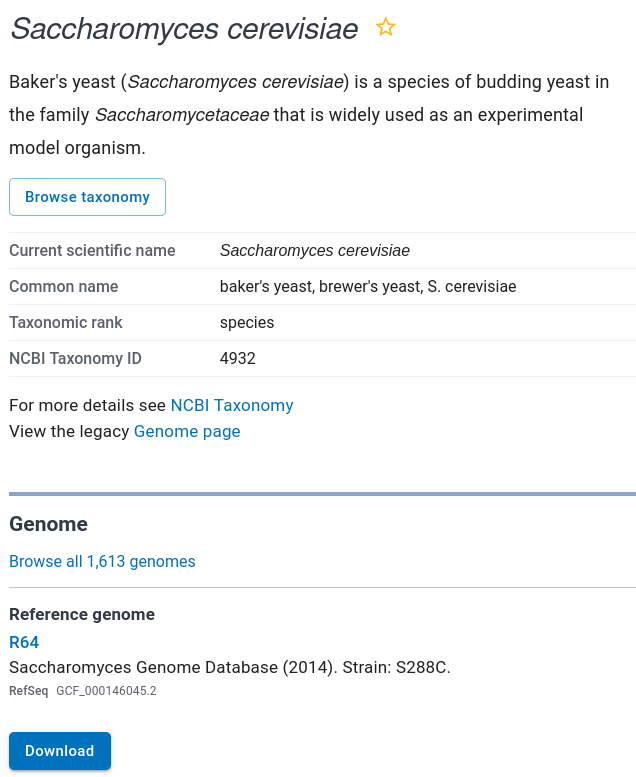
| Scroll down to the Reference genome and click
on "Download". This page has links for downloading sequences and annotation in a variety of formats
|
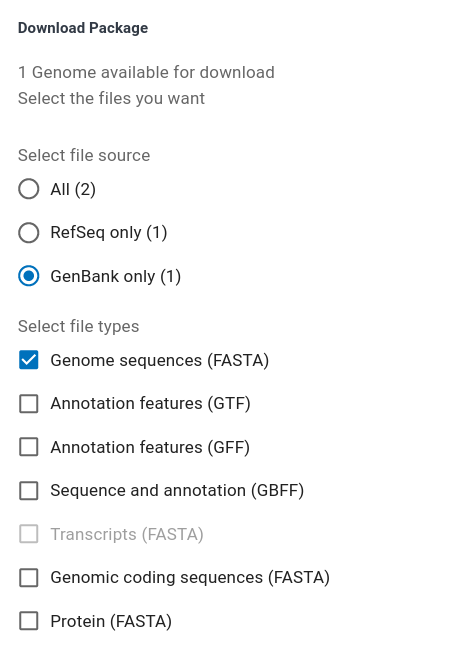 |
| Choose GenBank only and Genome sequences
(FASTA) Scroll down and click on the Download button 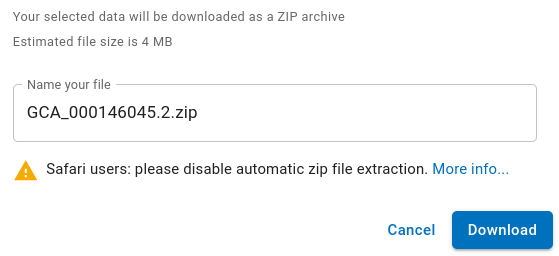 NOTE!!! Before downloading, make sure that your web browser is set to prompt you for a location to save your files. To keep things simple, it is best to avoid the Downloads directory, and save the file directly in the getgenome/temp directory. The next steps assume that the zip file is saved in temp. |
| If you need
annotation... It should be emphasized that FASTA
files do not contain any annotation information. For cases
in which you want to utilize this information, for example,
in browsing a genome, you would need to download either the
GFF file or the GenBank file. GenBank files contain both
annotation and sequence for each chromosome. |
There should now be a zip file:
-rw-r-----. 1 psgendb psgendb 3825906 Nov 9 19:09
GCA_000146045.2.zip
This file is an archive containing some metadata (information on
the files) as well as the FASTA sequence file itself. To
de-archive the file:
The directory will now look like this:
-rw-r-----. 1 psgendb psgendb 3825906
Nov 9 19:09 GCA_000146045.2.zip
drwxrwxr-x. 3 psgendb psgendb 4096
Nov 9 19:09 ncbi_dataset
-rw-------. 1 psgendb psgendb 1596
Nov 9 2023 README.md
Linux gives you some easy ways of finding out what is in a file.
Head - The head command lets you see the first few lines of a
file, just to have some idea of what is in it.
eg.
head GCA_000146045.2_R64_genomic.fna
will print the first 10 lines of this file
| >BK006935.2 TPA_inf: Saccharomyces
cerevisiae S288C chromosome I, complete sequence ccacaccacacccacacacccacacaccacaccacacaccacaccacacccacacacacacatCCTAACACTACCCTAAC ACAGCCCTAATCTAACCCTGGCCAACCTGTCTCTCAACTTACCCTCCATTACCCTGCCTCCACTCGTTACCCTGTCCCAT TCAACCATACCACTCCGAACCACCATCCATCCCTCTACTTACTACCACTCACCCACCGTTACCCTCCAATTACCCATATC CAACCCACTGCCACTTACCCTACCATTACCCTACCATCCACCATGACCTACTCACCATACTGTTCTTCTACCCACCATAT TGAAACGCTAACAAATGATCGTAAATAACACACACGTGCTTACCCTACCACTTTATACCACCACCACATGCCATACTCAC CCTCACTTGTATACTGATTTTACGTACGCACACGGATGCTACAGTATATACCATCTCAAACTTACCCTACTCTCAGATTC CACTTCACTCCATGGCCCATCTCTCACTGAATCAGTACCAAATGCACTCACATCATTATGCACGGCACTTGCCTCAGCGG TCTATACCCTGTGCCATTTACCCATAACGCCCATCATTATCCACATTTTGATATCTATATCTCATTCGGCGGTcccaaat attgtataaCTGCCCTTAATACATACGTTATACCACTTTTGCACCATATACTTACCACTCCATTTATATACACTTATGTC |
You could see a list of the sequences in this file by using the
grep program to search for lines containing the '>' character:
grep '>' GCA_000146045.2_R64_genomic.fna
produces the output
| >BK006935.2 TPA_inf: Saccharomyces
cerevisiae S288C chromosome I, complete sequence >BK006936.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome II, complete sequence >BK006937.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome III, complete sequence >BK006938.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome IV, complete sequence >BK006939.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome V, complete sequence >BK006940.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome VI, complete sequence >BK006941.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome VII, complete sequence >BK006934.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome VIII, complete sequence >BK006942.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome IX, complete sequence >BK006943.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome X, complete sequence >BK006944.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome XI, complete sequence >BK006945.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome XII, complete sequence >BK006946.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome XIII, complete sequence >BK006947.3 TPA_inf: Saccharomyces cerevisiae S288C chromosome XIV, complete sequence >BK006948.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome XV, complete sequence >BK006949.2 TPA_inf: Saccharomyces cerevisiae S288C chromosome XVI, complete sequence |
If you go back to the page listing Saccharomyces species, you can
find a link to S. arboricola. Click on
View in genome table.
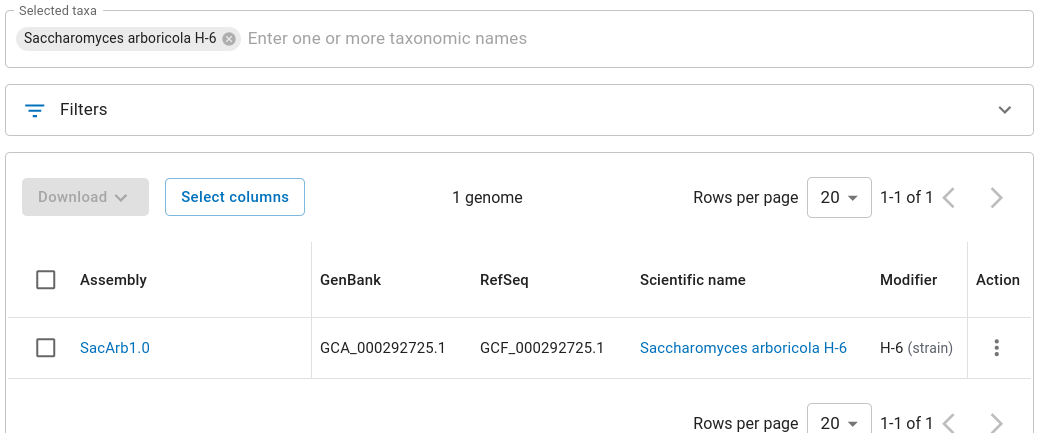
Choose SacArb1.0.
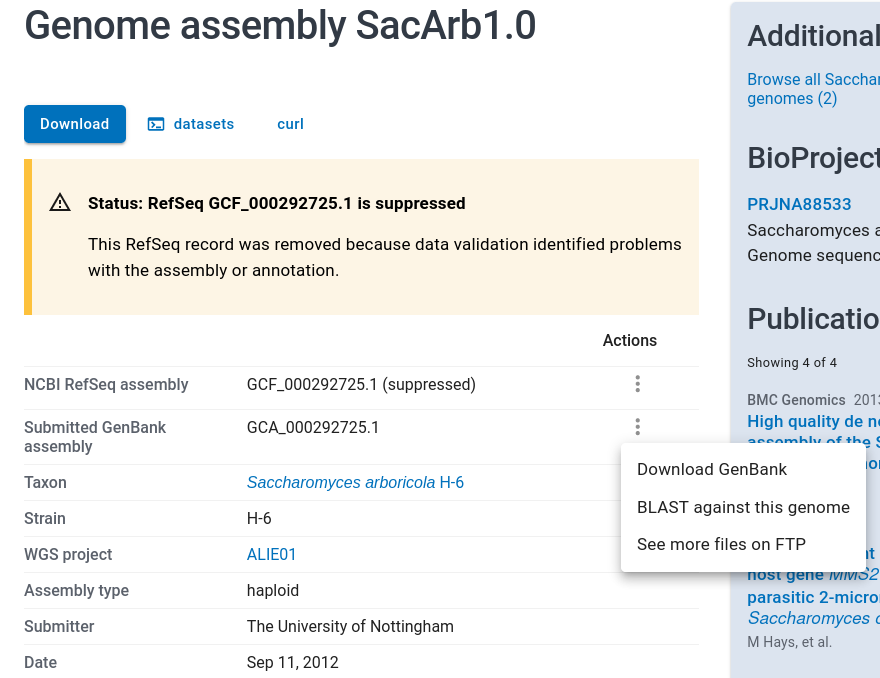
Open the three dot menu for "Submitted GenBank assembly", NOT for
the suppressed assembly. Click on Download. Once again, choose
GenBank only, and FASTA format. Download the zip file to your temp
directory, and extract the FASTA file as before.
Use the grep command to list the sequences in the fasta
file .
| >CM001563.1 Saccharomyces arboricola
H-6 chromosome I, whole genome shotgun sequence >CM001564.1 Saccharomyces arboricola H-6 chromosome II, whole genome shotgun sequence >CM001565.1 Saccharomyces arboricola H-6 chromosome III, whole genome shotgun sequence >CM001566.1 Saccharomyces arboricola H-6 chromosome IV, whole genome shotgun sequence >CM001567.1 Saccharomyces arboricola H-6 chromosome V, whole genome shotgun sequence >CM001568.1 Saccharomyces arboricola H-6 chromosome VI, whole genome shotgun sequence >CM001569.1 Saccharomyces arboricola H-6 chromosome VII, whole genome shotgun sequence >CM001570.1 Saccharomyces arboricola H-6 chromosome VIII, whole genome shotgun sequence >CM001571.1 Saccharomyces arboricola H-6 chromosome IX, whole genome shotgun sequence >CM001572.1 Saccharomyces arboricola H-6 chromosome X, whole genome shotgun sequence >CM001573.1 Saccharomyces arboricola H-6 chromosome XI, whole genome shotgun sequence >CM001574.1 Saccharomyces arboricola H-6 chromosome XII, whole genome shotgun sequence >CM001575.1 Saccharomyces arboricola H-6 chromosome XIII, whole genome shotgun sequence >CM001576.1 Saccharomyces arboricola H-6 chromosome XIV, whole genome shotgun sequence >CM001577.1 Saccharomyces arboricola H-6 chromosome XV, whole genome shotgun sequence >CM001578.1 Saccharomyces arboricola H-6 chromosome XVI, whole genome shotgun sequence >JH806614.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold1, whole genome shotgun sequence >JH806615.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold2, whole genome shotgun sequence >JH806616.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold3, whole genome shotgun sequence >JH806617.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold4, whole genome shotgun sequence >JH806618.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold5, whole genome shotgun sequence >JH806619.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold6, whole genome shotgun sequence >JH806620.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold7, whole genome shotgun sequence >JH806621.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold8, whole genome shotgun sequence >JH806622.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold9, whole genome shotgun sequence >JH806623.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold10, whole genome shotgun sequence >JH806624.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold11, whole genome shotgun sequence >JH806625.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold12, whole genome shotgun sequence >JH806626.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold13, whole genome shotgun sequence >JH806627.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold14, whole genome shotgun sequence >JH806628.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold15, whole genome shotgun sequence >JH806629.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold16, whole genome shotgun sequence >JH806630.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold17, whole genome shotgun sequence >JH806631.1 Saccharomyces arboricola H-6 unplaced genomic scaffold SU7_scaffold18, whole genome shotgun sequence >CM001579.1 Saccharomyces arboricola H-6 mitochondrion, whole genome shotgun sequence |
These genomes will be used in subsequent tutorials