CLVALID: Cluster Validation
CLVALID is technique for evaluating unsupervised clustering techniques.
Given the large number of clustering methods available to researchers, this module aims to evaluate the technique most appropriate for the data.
CLVALID uses the R package "clValid" to compare the relative properties of 10 different clustering methods across a several different numbers of clusters.
This module aims to help choose a method that is most compact, well-separated, connected, and stable.
It also makes use of bioconductor annotation packages to biologically validate the results.
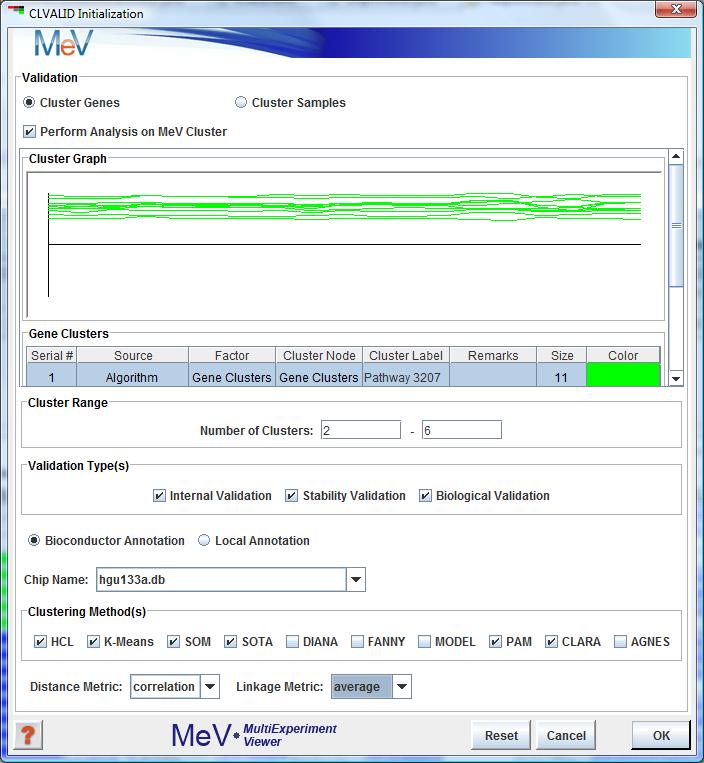 CLVALID initialization dialog box
CLVALID initialization dialog box
-The user must first choose whether the clustering validation is to occur over samples or genes. Please note that large datasets (typically over 10,000 genes) may result in memory issues.
If clustering a dataset with a large number of genes, you may instead choose to run the analysis on a smaller subset of your genes.
To do this, you must first create an MeV cluster using the Cluster Manager or other method.
-Once created, the "Perform Analysis on MeV Cluster" checkbox will be enabled. Click this and select the desired cluster using the Cluster Browser.
-Next, select the number of clusters involved in your analysis. The maximum cluster range is 2-6 clusters.
-Next, choose at least ONE type of validation. See below for a summary of validation types. If you are running biological validation, your data must contain an annotation field for "PROBE_ID" and you must select the appropriate Bioconductor annotation package from the drop-down menu.
-Finally, select at least ONE clustering method and distance metric. This module allows for the use of 10 different methods, of which HCL, KMC, SOTA and SOM are currently available for further exploration in MeV.
(If running HCL or AGNES, you must also select a linkage method.)
Parameters
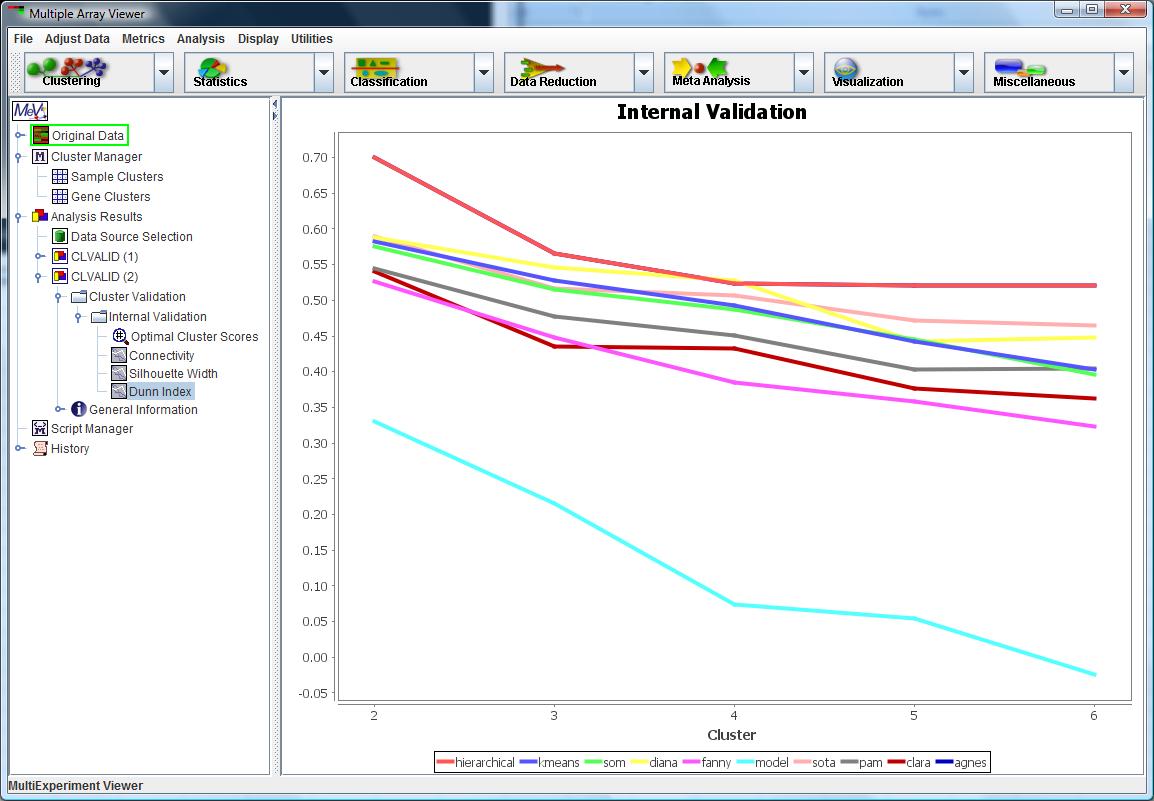 CLVALID Internal Validation Result
CLVALID Internal Validation Result
Validation Measures
Internal Validation: This validation evaluates the quality of the clustering based solely on the dataset and the clustering partition.
This assessment is demonstrated by the measures Connectivity, Silhouette Width and Dunn Index, which were chosen to elucidate the compactness,
connectedness and separation of the cluster partitions.
Stability Validation: This validation uses an iterative approach of removing one column (or row, if by genes) from the dataset and comparing the results.
Measures of stability include Average Proportion of Non-Overlap (APN), Average Distance (AD), Average Distance between Means(ADM) and Figure of Merit(FOM).
Biological Validation: This validation uses a bioconductor annotation package to find clusters with biological meaning. Measures include the Biological Homogeneity Index(BHI) and
Biological Stability Index(BSI).
(For more detailed information on validation options and available measures, see the R package vignette.)
Results
