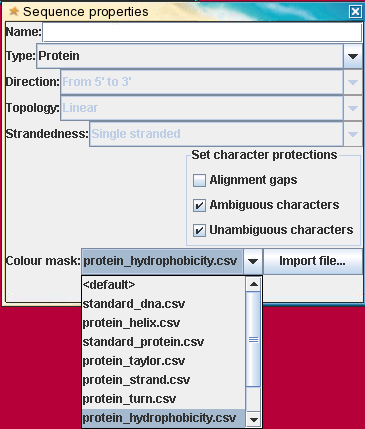
| Colourmask files -
bioLegato colourmask files assign a hexadecimal color code to sets of
characters. Any characters not listed in the file are displayed in
black. Files are two-column csv files, using the TAB character as the
seperator. The first line is a title line. On subsequent lines, a list
of one or more characters is included in the first field, and a
hexidecimal colour code is in the second field. For example the
contents of protein_zappo.csv are shown at right. |
aa colour
ILVAM #ffafaf
FWY #ffc800
KRH #6464ff
DE #ff0000
STNQ #00ff00
PG #ff00ff
C #ffff00
|