- Selecting datasets
- Overlaying data on a pathway
- Overlaying data on cellular overview
- Adding datasets
- Editing datasets
- Deleting datasets
-
Click config

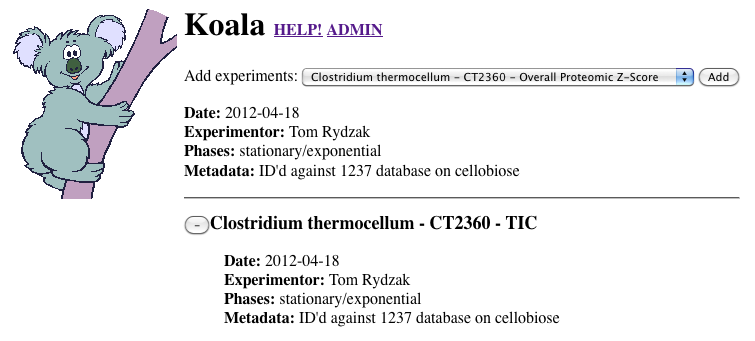
-
Click "-" to remove any unwanted datasets
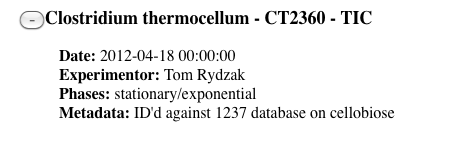
-
Select a dataset
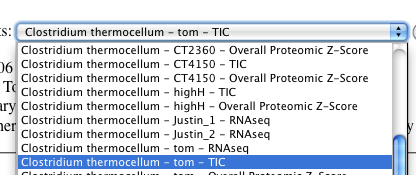

-
Click "add" to add the dataset


-
You are now ready to work with your data
-
Close the window and go back to the main pathway tools page

-
Refresh your browser, by clicking the refresh button


-
Now comes the fun part! You are ready to overlay your data on top of any page in biocyc!
Return to top
Warning: make sure you first have completed the above section "selecting datasets"
-
Refresh your browser, by clicking the refresh button


-
Change your organism to the desired organism by clicking "change organism databse"
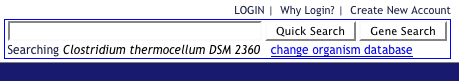
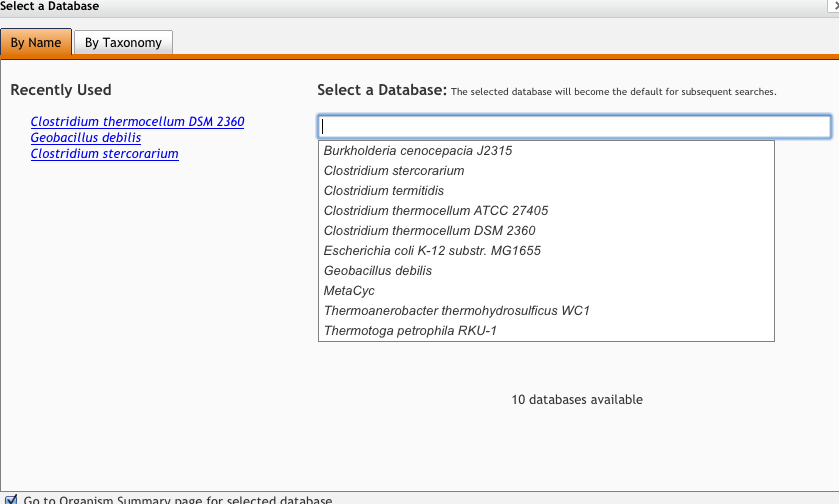

-
Enter a pathway in the search box and click "Quick search"

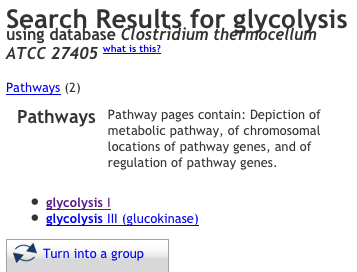
-
Select the pathway from the results you are interested in (in this example, we are interested in "superpathway of glycolysis, pyruvate dehydrogenase, TCA, and glyoxylate bypass"
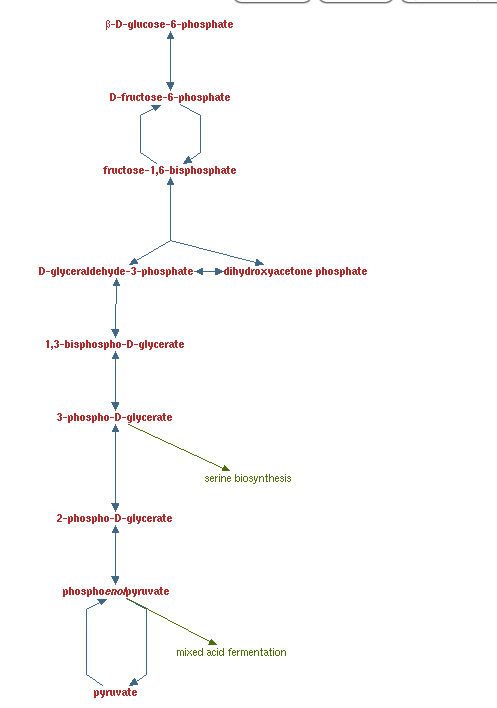
-
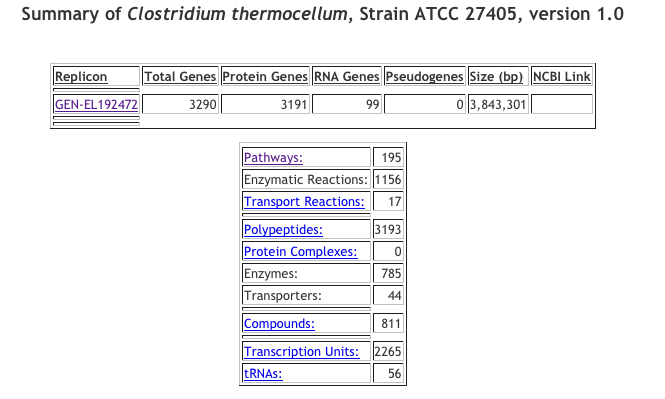
Click display


Return to top
Warning: make sure you first have completed the above section "selecting datasets"
-
Refresh your browser, by clicking the refresh button


-
Change your organism to the desired organism by clicking "change organism databse"






-
Open cellular overview (located under the tools menu)

-
Note: if you get the following screen, click "Dismiss" and retry this procedure (from the first step) in about 30 minutes or so (DO NOT TURN OFF THE COMPUTER!). If this message occurs more than once (i.e. 5+ times, for a total wait time of more than 150 minutes), please contact the pathway tools support staff for assistance.
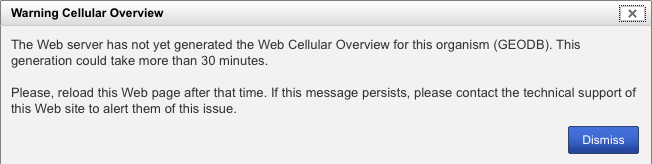
-
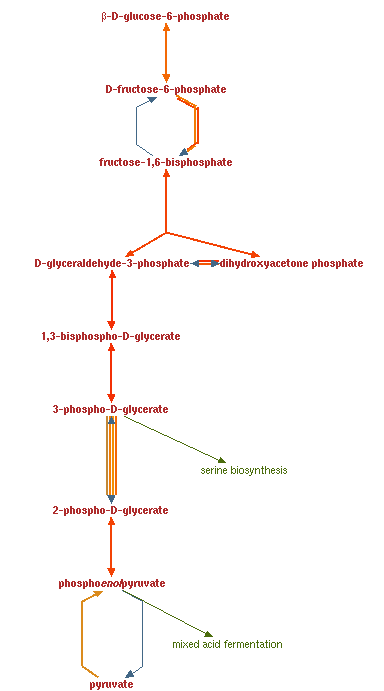
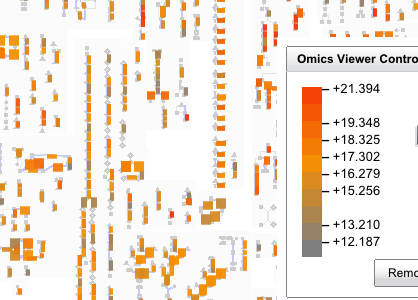
You should now see the cellular overview (blue arrows indicate that the enzyme is present in the current genome annotation)

-
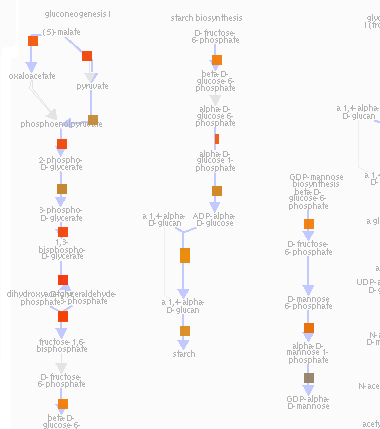
Click display (blue arrows are still present and still indicate that an enzyme is present in the current genome annotation)

-
NOTE: you can use the +/- and arrow controls to change the zoom and view of the pathways
Return to top
-
Click config


-
Click ADMIN

-
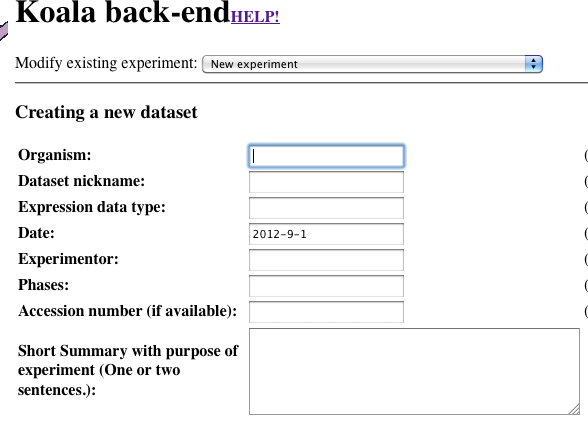
Ensure that the top dropdown says "New experiment"

-
Fill out the experiment data
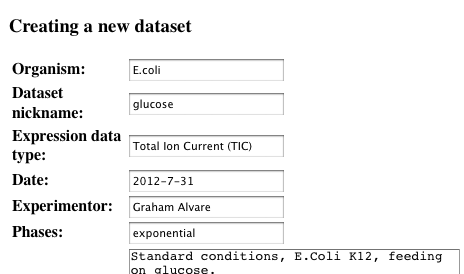
-
Select a dataset - the dataset must be contained in a TAB-delimited 2 column CSV file. The first column should contain gene identifiers, and the second column should contain the expression values, as shown below:
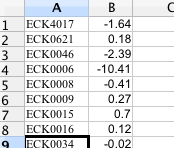
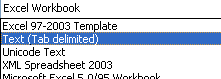
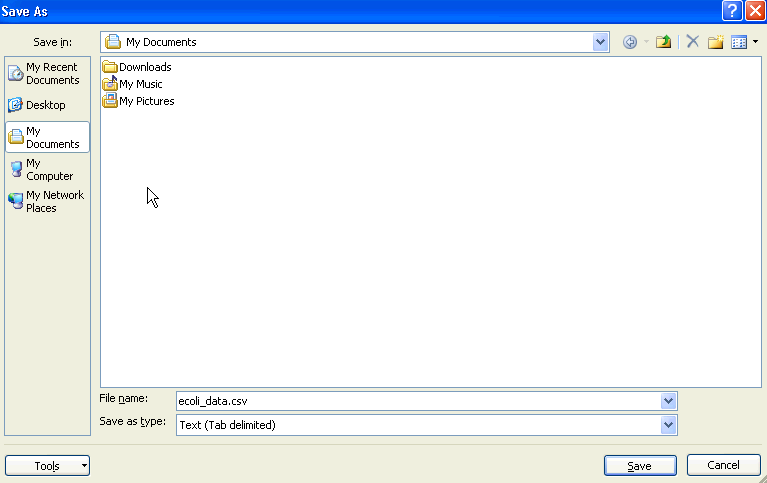
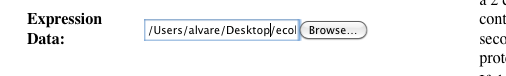
-
Determine whether the dataset should be scaled as log2, absolute scale, or both. If you are unsure, just ignore both of these checkboxes

-
Click add

Return to top
-
Click config


-
Click ADMIN


-
Select a dataset
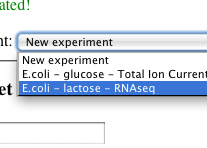
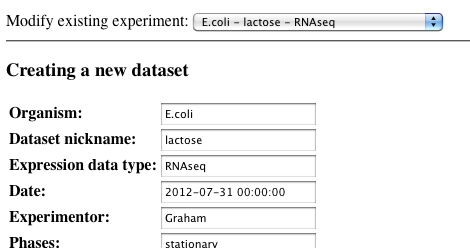
-
Make your changes to the dataset
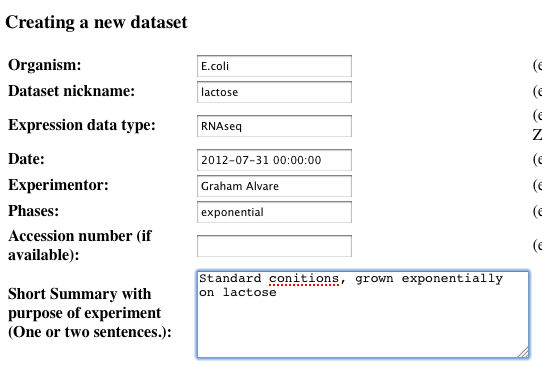
-
Click update


Return to top
-
Click config


-
Click ADMIN


-
Select a dataset
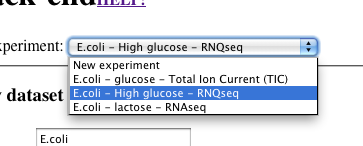
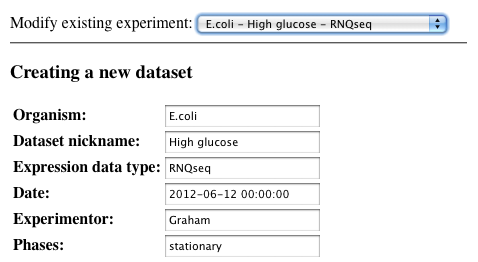
-
Verify that this is the dataset you wish to delete. If it is, then click delete.

-
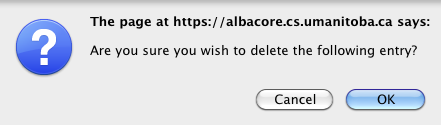
Confirm that you wish to delete.

Return to top















































































