SEQUENCE ASSEMBLY
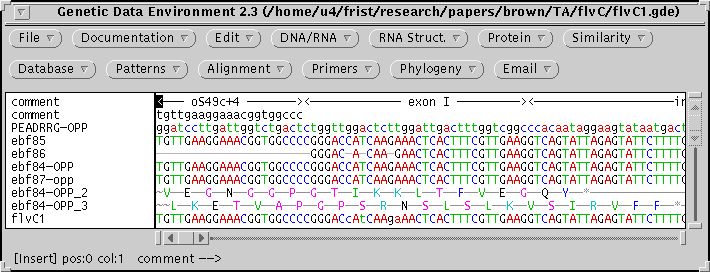
The figure illustrates the use of GDE for combining sequencing
reactions into a single contig.
- The raw sequence from each
sequencing reaction was read in from the File menu (ebf85 etc.).
- Flanking vector sequences were removed manually
- Only one strand of each reaction is needed. Where reactions are
opposite to the reading frame,
- sequence is selected
- DNA/RNA -> Complement to generate complementary strand
- Edit -> Reverse to flip sequence (restores 5'->3' orientation)
- File -> Get Info to add "-opp" to the name. This is a good way of
documenting the fact that the original reaction was on the other strand.
- To assist in assembling the sequence, comments are added in the top
two lines showing the locations of PCR primers and exon/intron boundaries.
Gaps are inserted by hand to align raw sequences.
- The composite sequence is generated by selecting all raw sequences and
choosing Alignment -> Sequence consensus.
- For verification purposes, the consensus is translated in 3 reading
frames, preserving codon spacing (DNA/RNA -> Translate). In the figure,
reading frames 2 & 3 are the reading frames of the 1st and 2nd exons.
Reading frame 1 was deleted from the alignment.
- The consensus is saved to a separate file using the File menu.
The final sequence file could be read using SEQUIN
to prepare a submission to GenBank.

