dgde &
Choose File --> Import namefile and read in temp.gi
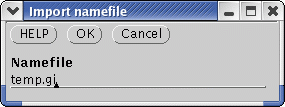
dgde & Choose File --> Import namefile and read in temp.gi |
 |
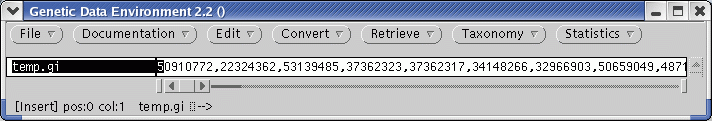
| Select the name temp.gi, and
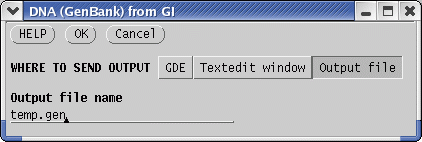
choose Retrieve --> DNA(GenBank)
from GI. Send output to a file called temp.gen. The GenBank entries will be written to temp.gen . This will be a temporary file, since the next step is to find other defensin genes by sequence similarity search. |
 |
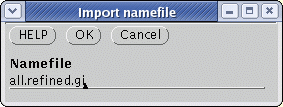
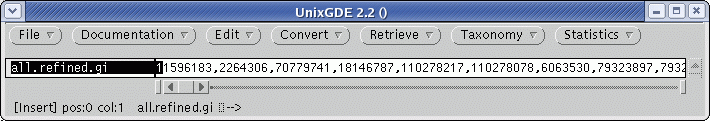
| Select
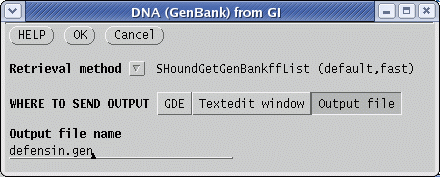
the .gi list as shown above, and choose Retrieve --> DNA
(GenBank)
from GI. Send the output to a file called defensin.gen. The file will be written to defensin.gen. |
 |