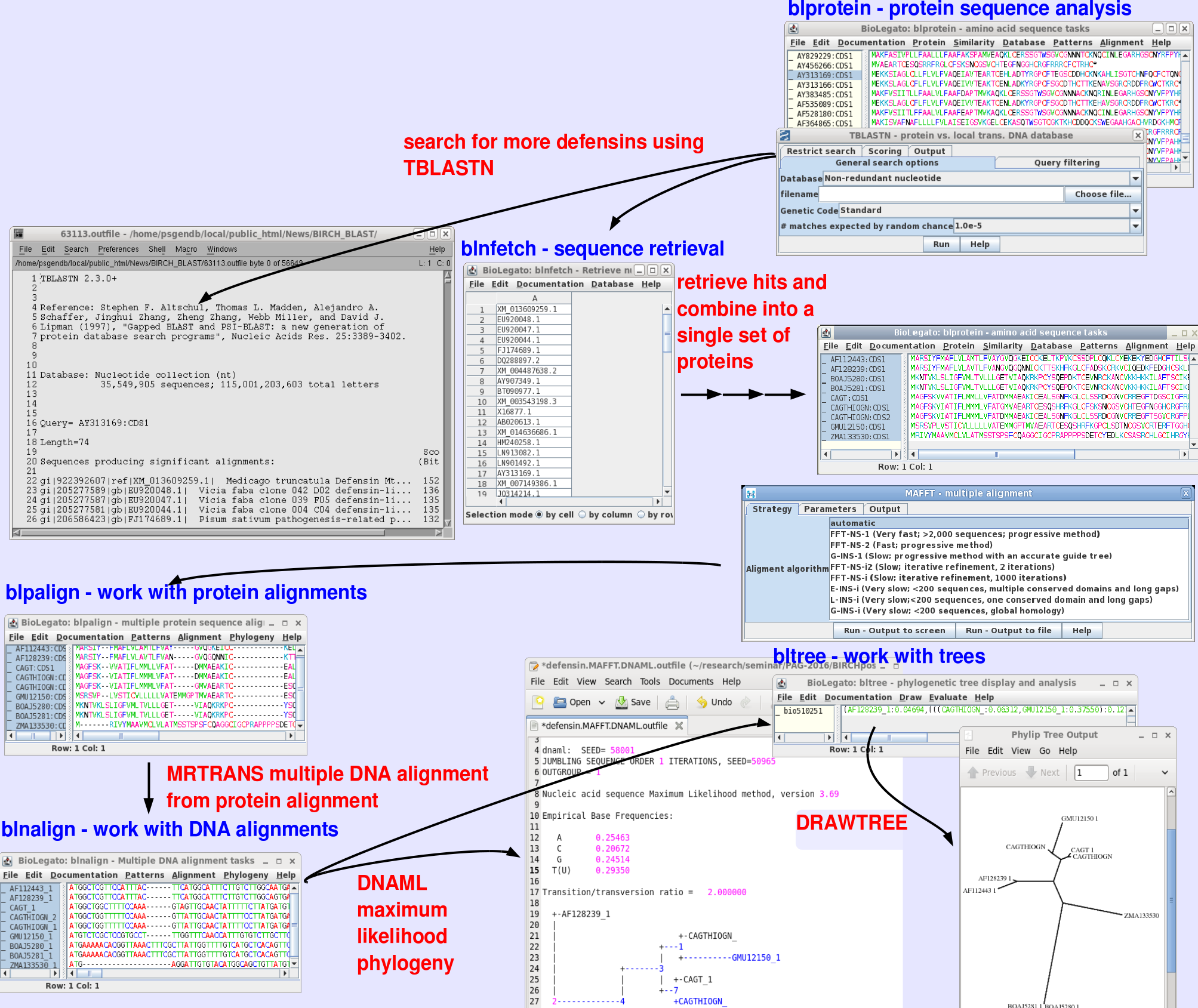
Installing and Searching
Local BLAST Databases
| |
Installing and Searching Local BLAST Databases |
May 8, 2016 |
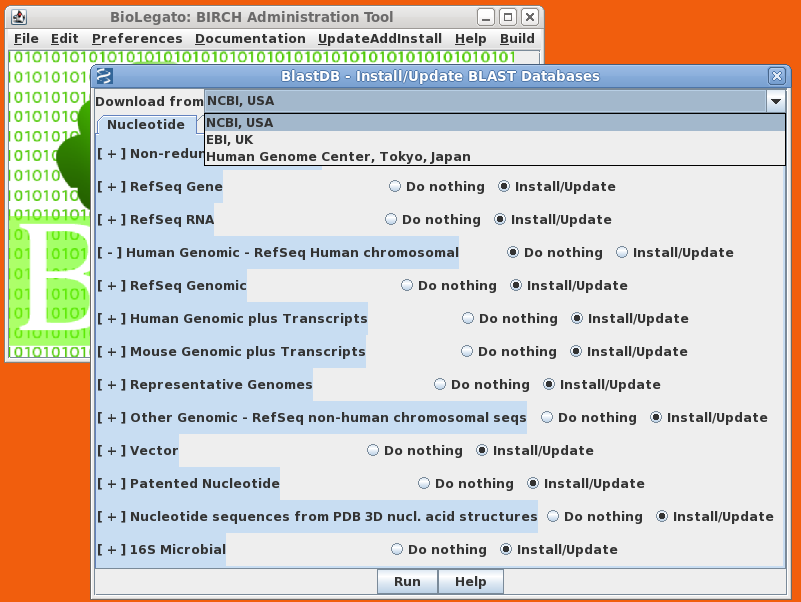
| The
BIRCH Administration Tool (birchadmin) makes it easy to
install, update and delete local copies of BLAST
databases. As illustrated at right, installation is as
simple as choosing the FTP site from which you wish to
download databases, and choosing the databases you wish to
install, update or delete. Since downloads of large
databases may take several hours, you can be notified by
email when the install is complete. To carry out these database management tasks, birchadmin calls the Python script blastdbkit.py, which can also be run at the command line. |
 |
 |
 |
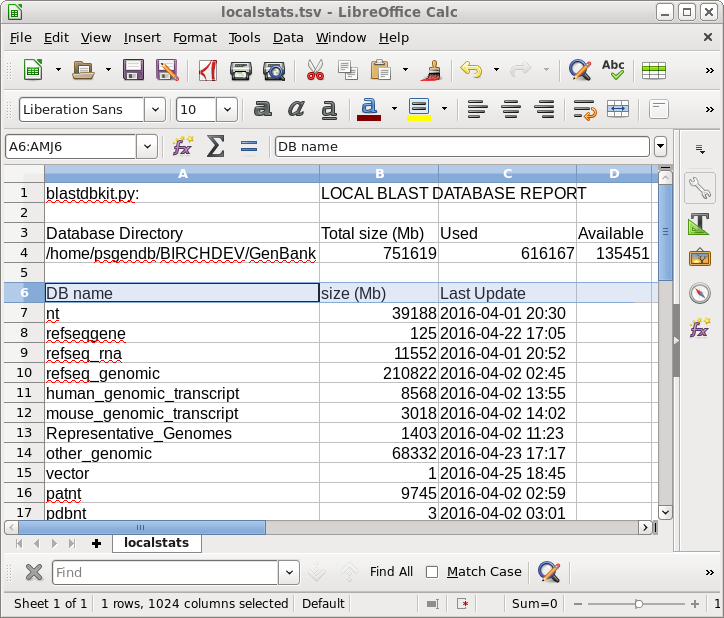
| Local
Database report |
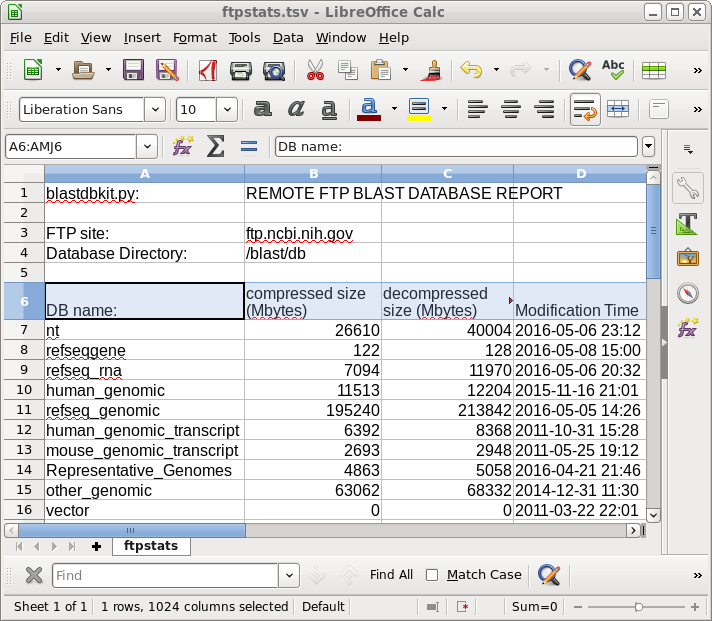
Report on
databases available for download |