TUTORIAL:
|
Oct. 21, 2014 |
TUTORIAL:
|
Oct. 21, 2014 |
This tutorial assumes that a local copy of the GenBank database is installed. |
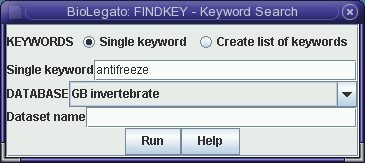
The FINDKEY Database menu allows you to select specific divisions of GenBank to search (eg. Primate, Rodent, Mammalian, Vertebrate etc.). Alternatively, FINDKEY can also search database subsets you have created yourself, containing GenBank entries.
FINDKEY does not retrieve sequence entires. Rather, it retrieves a hitfile and a namefile. The hitfile contains the lines that matched the keyword(s).
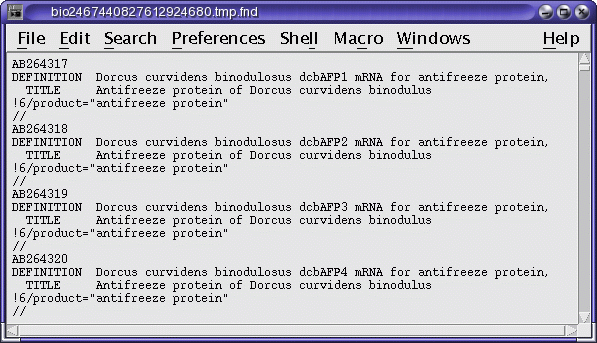
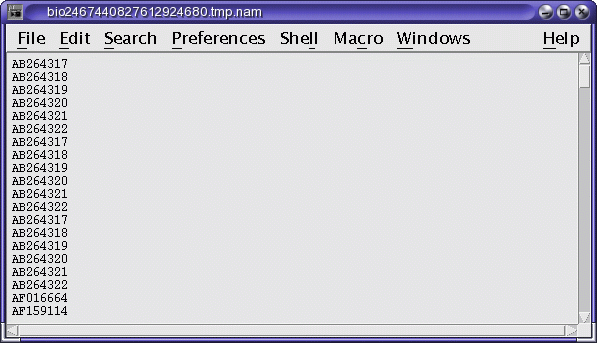
The namefile contains the names of the sequences found. These names can be directly copied and pasted to for retrieval by FETCH, as shown below. However, since the hitfile shows the hits in context, it is possible to eliminate some of the names from the list before retrieval.
There are several choices for WHERE TO SEND OUTPUT. Clicking on bldna would cause the sequences to appear in a new bldna window. This is convenient, but loses all the annotation in the GenBank entries. To retrieve the entries intact to a single file, choose "Textedit window" or "Output file". With the former, we have to wait for the entries to pop up in a textedit window. With the latter, the retrieval runs in the background. At this point you could even log out, and the retrieved file would be present in the directory in which bldna was run, the next time you logged in. If you choose "Output file" as in the example below, you must also type the name of a file to contain the output eg. "antifreeze.gen". Finally, FETCH can directly create XYLEM datasets, in which output is split into files containing annotation, sequence, and an index. See XYLEM documentation for details.
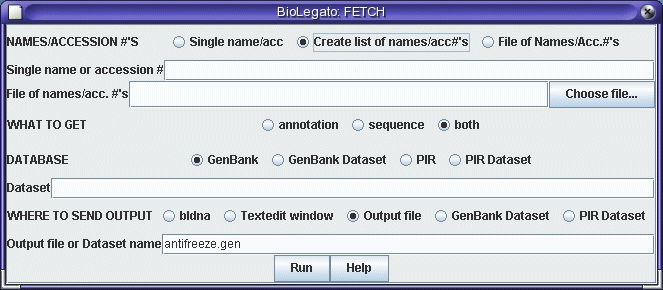
After you click "Run", a Text Edit window will pop up, into which you can paste the names or accession numbers of entries you wish to retrieve.
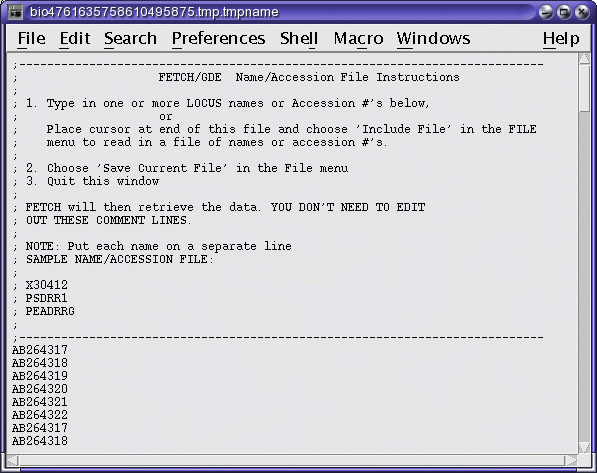
To begin the retrieval, choose File --> Save in the Text
Editor, and then quit the Editor. FETCH will
retrieve the GenBank entries and write them to the directory in
which
bldna
was launched. The retrieval may take several minutes. The message
"Fetch completed" will appear in the Terminal window from which
bldna was
launched.
There should then be a file called antifreeze.gen.