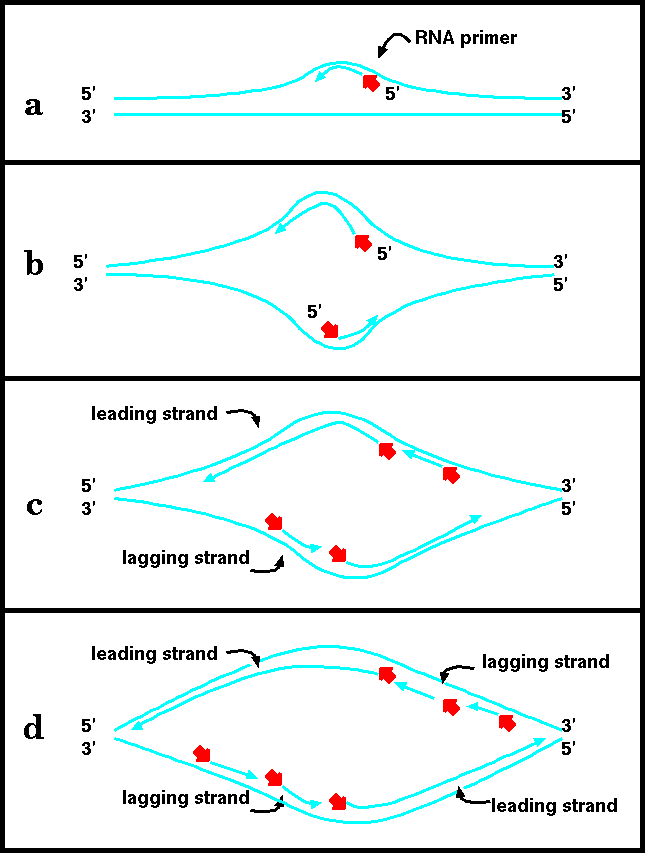
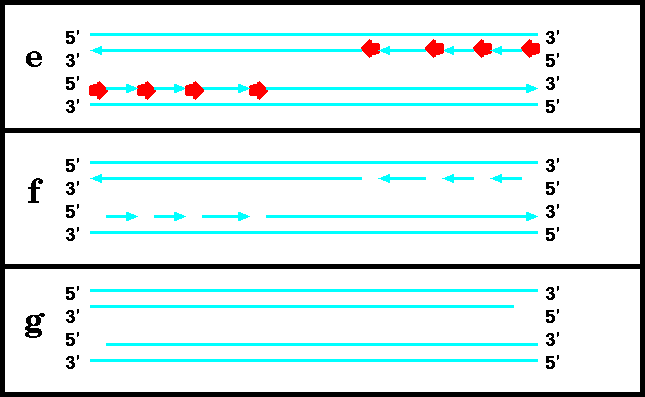
a) DNA polymerases extend a 3' end by adding nucleotides. DNA polymerases can only add nucleotides when there is a primer/template duplex.
DNA replication begins with the formation of short RNA primers, synthesized by primase. This is because DNA polymerases can not add nucleotides unless there is a pre-existing 3' end to add nucleotides to. Once the primer is in place, polymerase can elongate it. Finally, RNAase removes the primers, and DNA polymerase fills in the resulting gaps by extending the growing strand that is upstream. When the last nucleotide is filled in, the resultant gap is closed by ligase.
b) As the helix continues to open up, enough room is available to allow synthesis of a primer, and its subsequent elongation, on the opposite strand.
c) Elongation of the leading strands proceeds both leftward and rightward. The leading strands undergo continuous elongation. As the helix opens up, new primers can be added to the lagging strands, and the primers elongated by DNA polymerase.
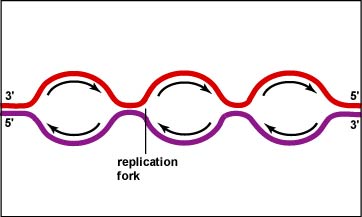
d) Each leading/lagging strand pair constitutes a "replication fork". Thus, the replication forks migrate leftward and rightward, opening up the helix and allowing for new primers to be inserted on the lagging strands.