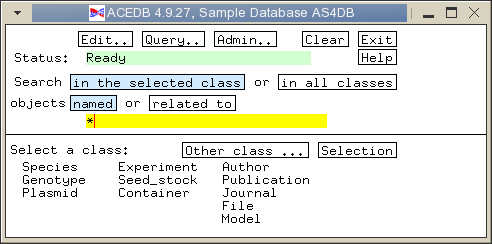
For every class that you add to models.wrm, you must also add a line to wspec/options.wrm, indicating whether it is a visible class ie. whether it should be displayed in the main window. For example, to add the DNA_sample class, the following line would be added to options.wrm:
_VDNA_sample -V
(Both the _V and -V are required. More detailed instructions are included in the options.wrm file.
To read in the updated models.wrm file, go to the main ACEDB window and choose Edit--> Read models. If there are no errors in models.wrm, no error messages will be reported. If there are errors, an window will pop up telling you which model has an error, and on which line the error occurs.
Next, you have to tell ACEDB to display the DNA_sample class in the main window. Click on the 'Selection' button in the main window, and the following window will pop up:
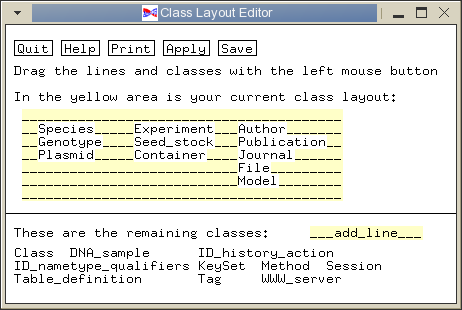
Use the left mouse button to drag the text 'DNA_sample' into the yellow class layout block. Place DNA_sample between Seed_stock and Container, and click Save.
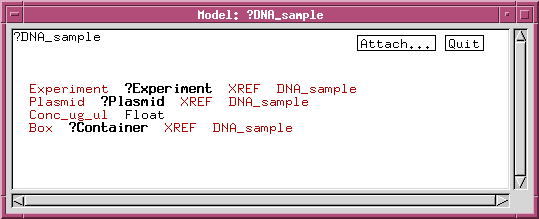
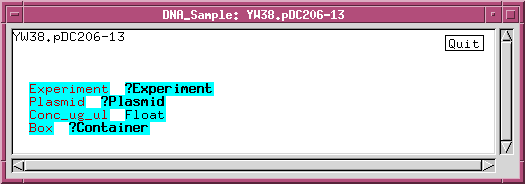
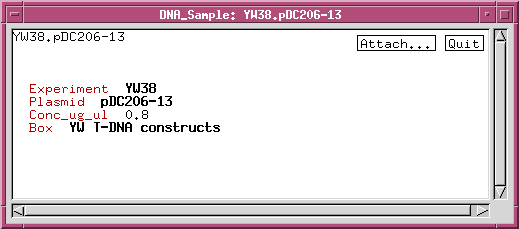
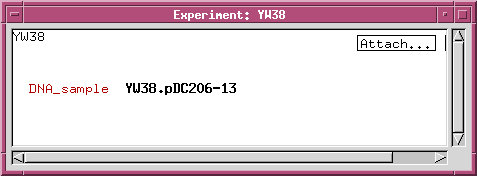
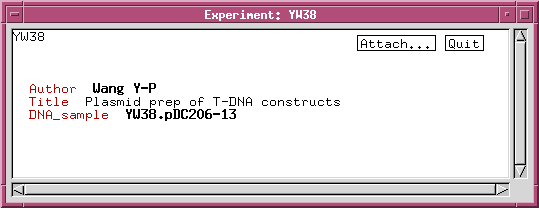
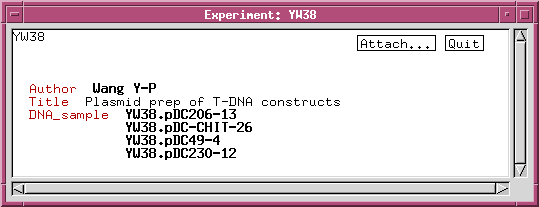
We can refine the DNA_sample class by adding data fields or links to other classes. For example