Research
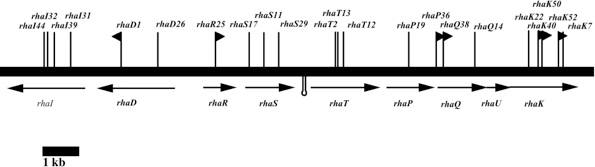
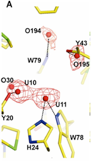
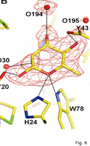
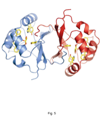
Characterization of Rhamnose catabolism in Rhizobium. Initial interest in rhamnose catabolism arose from the observation that it was necessary for competition for nodulation (Oresnik et al 1998). To elucidate the role of rhamnose catabolism the locus that complements the inability to utilize rhamnose was characterized genetically, biochemically and physiologically (Richardson et al 2004). The work suggested that the rhamnose catabolic pathway was atypical; whereas E. coli initiates the catabolism or rhamnose with an isomerization, R. leguminosarum initiates catabolism by a phosphorylation. It was subsequently shown that not only did the kinase (RhaK) act directly on rhamnose, it also affected the transport of the sugar (Richarson and Oresnik 2007). In addition, we have also able to crystallize RhaU, a small protein encoded within the operon that contained RhaK and an ABC transporter for rhamnose. The data showed that RhaU is a mutarotase and that its substrate was also rhamnose and not a phosphorylated form of the sugar (Richardson et al 2008). Interestingly this suggests that the kinase activity and the transport phenotype associated with RhaK can be uncoupled. Linker scanning mutagenesis of rhaK has yielded 2 mutants in which transport and kinase activities are uncoupled in RhaK. Our current work is focusing on characterizing these variant forms of RhaK. This work is funded by an NSERC Discovery grant.
Rhamnose locus; RhaU without and with rhamnose; ribbon diagram of RhaU dimer.
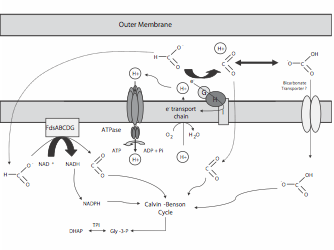
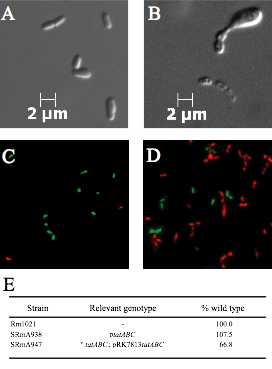
Characterization of Periplasmic redox proteins in Rhizobium. Our objectives are twofold. The first is to test the hypothesis that periplasmic redox systems are important to the physiology of Rhizobium in the soil. Since a number of periplasmic redox proteins utilize the twin arginine transport (tat) system, we initiated a characterization of the S. meliloti tatABC genes (Pickering and Oresnik 2010). As well our work led to the characterization of the tat-leader containing formate dehydrogenase FdoGHI where we were able to show S. meliloti is capable of formate dependent autotrophic growth (Pickering and Oresnik 2008). Our current work is focused on systematically deleting a number of putative tat-leader containing genes to determine if they have a role in the symbiotic process. This work is funded by an NSERC Discovery Grant.
Live/Dead images of S. meliloti suggesting that loss of tatABC is correlated with cell dysfunction; a metabolic scheme outlining the role of formate dehydrogenases and their role in autotrophic growth.
What role does Magnesium play in the physiology of S. meliloti? The ability to regulate genes to respond to environmental conditions is an essential biological characteristic. An understanding of the genetic determinants involved in any response will inevitably allow the identification of the pertinent biochemical and physiological parameters of a biological system. We have observed that some cations (magnesium and potassium) alter the morphology of S. meliloti (Miller-Williams et al 2006).
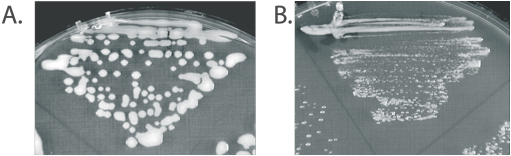
Mucoidy is ion dependent. LB agar plates of an expR+ S. meliloti grown without (A) or with a supplement of 350 mM KCl.
Our objective is to identify and characterize determinants that play a role in allowing S. meliloti to adjust to different ion concentrations. We have pursued this by isolating Tn5 mutants that no longer have a differential colony phenotype when grown on higher concentrations of Mg2+ or K+. This work is funded by an NSERC Discovery Grant.
Quantification of B. japonicum in Field Soils using a PCR Based Diagnostic Assay and the Relationship of B. japonicum to other Rhizobacteria found in the Field. The successful use of Bradyrhizobium japonicum as an inoculant on soybean is dependent upon the number of effective bacteria that are present as well as the ability of the bacteria to colonize the soil and to interact with the host plant. This proposal sets out to accomplish 2 major objectives. The first is to complete the optimization of a PCR based assay (culture independent) to enable the quantification of B. japonicum in soil samples in a matter of days. The second part of this proposal initiates a program of study using this assay, with other culture independent approaches, to determine how and where B. japonicum populations develop following inoculation onto a field. It is hoped that this information will be useful in developing better commercial inoculums. This work is funded by a research grant from the Manitoba Pulse Growers Association and Manitoba Agriculture.