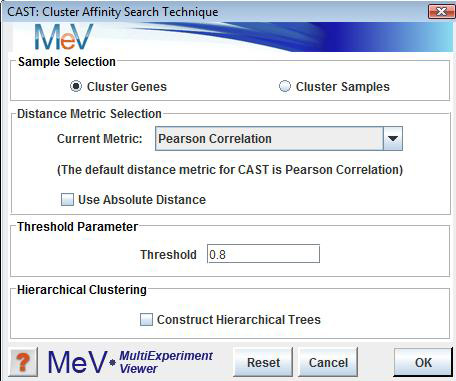
CAST Initialization Dialog
(Ben-Dor et al. 1999)
The user is prompted for a threshold affinity value between 0 and 1 (which may be thought of as the reciprocal of the distance metric between two genes, scaled between 0 and 1), that has to be exceeded by all genes within a cluster. The algorithm works by both adding and removing genes from a cluster, each time adjusting the affinities of the genes to the current cluster, and continuing this process until no further changes can be made to the current cluster.
Sample Selection
The sample selection option indicates whether to cluster genes or samples.
Threshold
The threshold parameter is a value ranging from 0.0 to 1.0 which is used as a cluster affinity threshold. Each expression element will have an affinity for the current cluster being created based on it's relationship to the elements currently in the cluster. If that affinity is greater than the supplied threshold the gene is permitted to be a member of the cluster. Note that thresholds near 1.0 are more stringent and tend to produce many clusters with rather low variability.
Conversely, using a lower threshold will produce fewer, more variable clustersA balance by trail-and-error should be found between these extremes.Hierarchical Clustering
This check box selects whether to perform hierarchical clustering on the elements in each cluster created.
Default Distance Metric: Euclidean