Button Selection
Global ANCOVA, a new module to MeV 4.6, is a technique for identifying differentially expressed gene sets based off of the calculation of an F-test between groups of samples. Analyses are typically run in a two-class format but may also be applied to additional groups, time-course or other designs of arbitrary variables. Global Ancova fits linear models to the data and compares them using the extra sum of squares principle. The result table reports p-values, permutation p-values and asymptotic p-values.
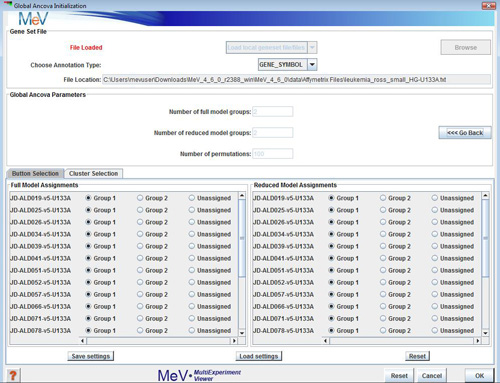
After opening the Global Ancova initialization dialog, select the file location of your gene set test terms. Your options are:
Choose an annotation type. The annotation used by your test terms must match those selected here.
Assign your samples
Choose the number of groups in your data and use the button or cluster selector to assign the samples to those groups.
Button Selection
Use this option if you have not created clusters for your samples and want to assign each sample individually.
Cluster Selection
Use the cluster selection panel if you have previously made clusters and wish to run your analysis based on those clusters. Use the drop-down boxes to choose which clusters’ samples are to be assigned to which time-points. For clusters you are not using, leave “unassigned”. The same condition and time-point requirements exist for this method, but the cluster selector makes for easier and more organized sample assignment.
The Global Ancova module outputs a table containg F-values, permutation-based p-values and approximation-based p-values.
Additionally, the Global Ancova module outputs standard viewers for each of the supplied genesets.