New Script Attribute Dialog
The scripting capabilities within MeV permit the execution of multiple algorithms to be performed without user oversight or intervention once processing begins. The execution steps are dictated by a user-defined script that describes the parameters to use for the selected algorithms. Scripting in MeV allows one to document the algorithms run and the selected parameters during data analysis. The script document can be shared with collaborators so that analysis steps can be replicated on the common data set. Scripting is also useful when running several long analysis steps that would normally require monitoring in MeV's interactive mode. Each algorithm and the parameters are pre-selected in the script so the next algorithm kicks off as soon as the previous run finishes. Despite the advantages of scripting, there may be times when careful evaluation of a result before deciding on the next algorithms is needed. In this setting scripting might be used as a first pass analysis and the multiple results of the script run can lead to the selection of different algorithms or new parameter selections.
The MeV script is an XML based text document containing information about which algorithms to run, the order of the algorithms, and the source data for each algorithm. Script creation is accomplished through a graphical representation of the script to eliminate the need for the user to understand the complex structure of the script. The Document Type Definition (DTD) can be found in Section 14 Appendix (MeV’s Script DTD) for those interested in the details of script structure.
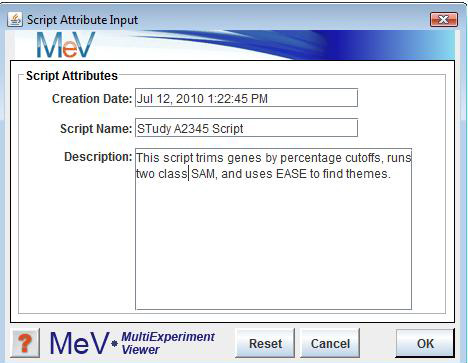
Creating a script is a simple process that can be initiated by selecting New Script from the menu in the Multiple Array Viewer. Data must be loaded before this menu option can be enabled since many algorithms require data-specific information (e.g. group assignments for TTEST or SAM depend on the number and order of the loaded experiments). Once the New Script menu option has been selected, an initial dialog form will come up to allow one to enter a script name and description.
The script name, description, and the creation date will be stored in the script as comments. Once the initialization dialog is dismissed the script manager node will become populated with a script table and two viewers associated with the new script. The viewer that opens automatically is called the . This viewer is a graphical representation of the script and it is from this viewer that the user constructs the script. The other script viewer is the , which displays the actual text of the script during script creation.
The Script Tree Viewer is the main viewer used to construct the script. The viewer's graphical nature permits the user to focus on script creation without undue consideration of complex script syntax. The Script Tree Viewer represents the script as a set of connected nodes. Each node is either a or an .
Data nodes, shown as light green, are sources for data for attached algorithms. Any number of algorithms can be attached to a data node. Algorithm nodes, shown as light yellow, represent processes that transform the data or act on the data to produce results.Select a data node to use as source data by left clicking a data node. Selected nodes will have a blue highlighted border when selected. A right click will reveal a menu containing an menu option. Add Algorithm Node will present a dialog used to select the algorithm and parameters to append to the data node.
The algorithms fall into three main categories represented on three tabbed panels in the Algorithm Selection Dialog. Analysis Algorithms include gene and experiment clustering algorithms, classification algorithms, statistical algorithms, and data visualizations. The analysis algorithms are all described in the section of the manual. The Adjustment algorithms are those algorithms found in the Adjustment menu of the Multiple Array Viewer interface and include the Affymetrix™ based filters if Affymetrix ™ data is currently loaded. The Adjustment algorithms either filter the data based on some criteria or are used to perform a mathematical transformation of the data.
Two main options are available for cluster selection. The output nodes from cluster selection on the Script Tree can be used as input data to new algorithms.
Diversity Ranking Cluster Selection computes cluster diversity for each of the input clusters and then ranks the clusters from least variable to most variable. Clusters are selected that satisfy a minimum size (population) but are as least variable as possible.The other option for cluster selection is Centroid Entropy/Variance Ranking Cluster Selection. This method places either a variance or an entropy value on the cluster’s (mean expression pattern).
Right click menus displayed from the script tree viewers vary depending on whether the selected node is a data node or an algorithm node. The menu displayed from a data node provides the and options. The menu displayed from an algorithm node provides the and options. The Delete Algorithm option deletes the algorithm, the associated output data nodes and any downstream algorithms that rely on the output of the deleted algorithm. The View XML Section option is a shortcut to the When selected this option will open the XML viewer and will highlight the algorithm section associated with the node selected in the Script Tree Viewer.
The Script XML Viewer is a text rendering of the script, as it would appear when saved to an output file. The main purpose of the viewer is to get a view of the script during script creation and to review parameter value selections for particular algorithms. When the XML viewer is opened via the Script Tree Viewer the selected algorithm in the XML viewer is highlighted in light green as shown in the Script XML Viewer figure. Script lines are selected by clicking on the row number displayed on the left side of the viewer. If the selected line corresponds to a parameter key-value pair, then the Edit menu option will be enabled so that the value can be altered.
Limited editing capabilities are available in the Script XML Viewer. If the selected script line is a parameter line then hitting the menu option will display an input dialog to permit altering the value of the parameter.
Script Value Editor
The Script Value Editor is used to make controlled changes to the script parameters. If a line in the XMLViewer contains a key:value pair then the Edit menu option is enabled. The entered value is validated for type and that it falls within possible magnitude constraints. If an error has occurred then the update will be halted and a warning will be issued.
The View Valid Parameters button will provide a table of parameters names, their types, possible constraints, and if the parameter is strictly required in all cases.
Great care should be taken to insure that changes to script parameters will not affect other algorithm parameters or the nature of the output such that down-stream algorithms are still appropriate.
Keep in mind that caution should be used when changing script parameters so that downstream algorithms that are dependent on results are still valid. The parameter input dialog only permits alteration of the parameter’s value. A button on the dialog, labeled will produce a table of possible parameters for the algorithm being modified. The list contains parameter names, parameter types, optional constraints (value limits), and whether the parameter is required in all cases or if it is dependent on other parameter selections.Loading of saved scripts is done by selecting from the File menu of the Multiple Array Viewer. A file selection dialog will automatically launch to prompt for the selection of a script file. Scripts should be stored in the script directory of MeV’s data directory to ensure proper validation.
During script loading several levels of script validation occur. (usually malformed XML), (script that does not match the Document Type Description (DTD)), , and algorithm (missing required parameters, parameter type mismatch, or parameter out-of-bounds errors) are caught during the validation. If multiple validation errors exist, all will be reported. All validation errors are reported in a Script Error Log dialog. The Error Log initially lists the errors and indicates a line number for each error. The button launches an XML viewer that can be modified and saved to address the errors. Once the dialog is closed, the script should be re-loaded using the File menu to begin a fresh script loading and validation.
The Script Error Log records and reports errors during script validation.
These errors fall into two basic categories.
Parse Errors describes problems with script construction and can be described as Fatal Errors
(malformed XML) such as missing closing tags, Validation Errors in which the script fails
to conform the the DTD (Document Type Description), and Parse Warnings.
Aside from Parse Errors, the other major error type is in parameter validation, Parameter Errors. These errors report missing parameters, parameter type mismatches, and parameters that fall outside of established range constraints.
The Edit Script button will display the current script in an editor window to the right of the reported error tables. If corrections are made to the script it is possible to right click to save the corrected script to file. If the validation error was reported during a script load then the load will be terminated but can be retried after script editing.
Script execution can be initiated from the Script Table viewer, the Script Tree Viewer, or the Script XML Viewer by using the right click menu option, . The script is logically split up into units called that represent one or more algorithms sharing a common input data source. The output results are grouped into algorithm sets and since each algorithm set has a unique input data set that can be a subset of the loaded data, an input data node is used to display the input data for the algorithm set. The figure displaying script analysis output clearly shows the expected output from a script containing one algorithm (SAM). The node shows the number of experiments and the number of genes as well as three cluster viewers. The node in the output for an algorithm set helps to orient the researcher as to which part of the script falls within the enclosing algorithm set. The algorithm set, input data, attached algorithms and the result data nodes, are highlighted while other script nodes are semi transparent.
If an algorithm fails to produces a data node that is a source data node for another algorithm set then that algorithm set (using the null input) is aborted and an empty node with a text label indicating empty source data is displayed.