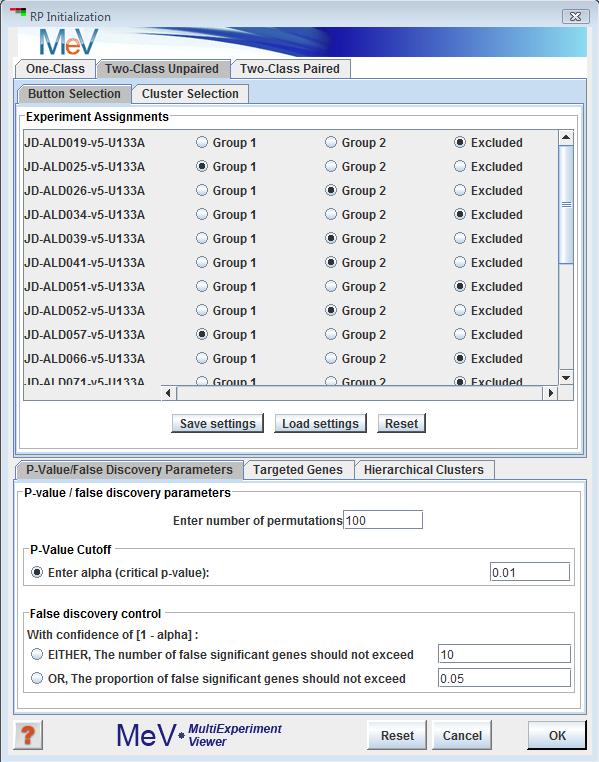
Rank Product Initilalization Dialog
(Breitling, Rainer et al, 2004)
Rank Products is a novel test for determining differential expressed genes with multiple replicates. This analysis differs from many other techniques in that it does not apply a sophisticated statistical model, but rather from the calculation of rank products, a faster and simpler method.
Additionally, Rank Products is useful in highly noisy data and can significantly reduce the number of replicate experiments required to obtain reliable results.
MeV’s RP currently supports 3 experimental designs.
As in most other modules, MeV offers two forms of sample selection: the individual button selectionthe cluster selection tabs to assign your samples to the analysis. Samples left unassigned or unchecked will be ignored in the analysis.
P-Value/ False Discovery Parameters
For determination of significance levels, specify the number of random permutations you want RP to run. If setting a significance cut-off using p-values, enter the alpha value you wish to set as the cut-off point. If determining significance by false discovery rate, check the box next to either the number or percentage of false positives and input the corresponding value.
Targeted Genes
Check the radio button for determining in the analysis significantly up-regulate genes, down-regulated genes or both.
Hierarchical Clustering
To have RP construct hierarchical trees for your results, check the corresponding check box. Select whether this feature is to be applied to significant genes or significant and non-significant genes. This process may add significantly to the computation time.
The RP module outputs standard viewers and tables for MeV’s statistics analyses.