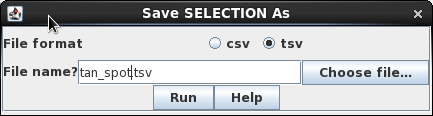
The results from any blncbi window can be saved by choosing Edit --> Select All, and then choosing File --> Save SELECTION As.
Make sure that the File format is tsv, and give the file a descriptive name with the .tsv file extension.
Due by 11:59 pm, Monday February 24.
| Search tips: 1) blncbi lets you construct complex queries using the conjunctions AND, OR and NOT, as well as grouping two or more things using parentheses. For example, a search aimed at retrieving either the pUC18 or pUC19 vectors might use the query (pUC18 [Title] OR pUC19 [Title]) AND syn [Division] 2) Don't waste your time adding trivial terms to your query to specifically exclude sequences by Accession number. eg. (pUC18 [Title] OR pUC19 [Title]) AND syn [Division] NOT (S38358 [Accession] OR M22135 [Accession] OR X13074 [Accession] OR X13070 [Accession]) If the number of false positives is small, they will be easy to weed out by inspection of the GenBank files. 3) Whole genome shotgun sequencing contigs and scaffolds may show up as hits. Although these probably do contain the gene you're looking for, it adds a layer of complexity to work with contigs, because they will contain many genes. Ideally, we want to find a smaller GenBank sequence that has a single tan spot gene, and no other genes. |
| Saving blncbi results The results from any blncbi window can be saved by choosing Edit --> Select All, and then choosing File --> Save SELECTION As. Make sure that the File format is tsv, and give the file a descriptive name with the .tsv file extension. |
 |
| TSV
stands for "Tab-separated value" files. This is a
generic format in which each row in a table has one or
more fields (columns). The values on each line are
separated by TAB characters, which have the same effect
as using tabs in a document. Virtually all spreadsheet
programs such as LibreOffice of MS Excell can import TSV
files. |
|
| Check your files It is always a good idea to examine your GenBank or TSV files in a text editor to make sure that they contain what you think they contain! |