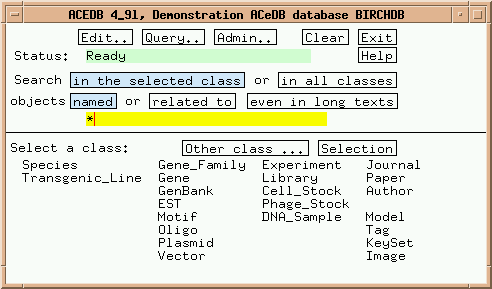
Now let's get a feeling for how ACEDB databases work. Click on "Gene_Family" in the main window, and the Main Keyset should appear as shown:
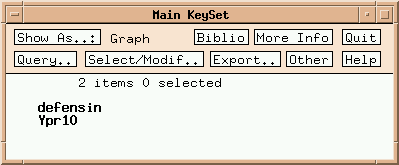
Two multigene families are listed. Click on 'defensin':
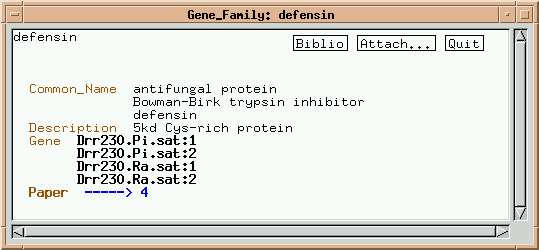
Click on "Drr230.Pi.sat:1":
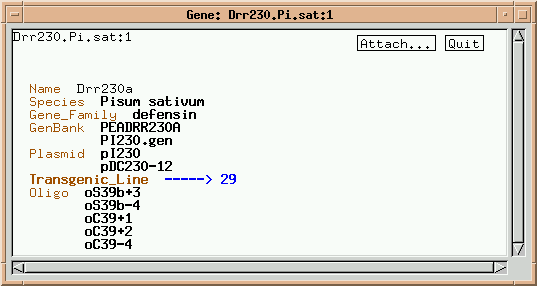
Note that this gene has links to some "higher order" classes, such as Gene_Family and "Species", as well as links to "lower order" classes, such as Plasmid and Oligo. Two plasmids containing this member of the defensin multigene family are listed. Click on pI230.
To view the map, click on "pI230.gif"
Note that this window contians the $ACE_VIEW_COMMAND environment variable created by the ACErunexternal. While we could have simply put in 'xv' in its place, it we later wanted to switch to a different graphics viewer, or if the xv command was moved to a new directory, all members of the Image class would have to be dumped into a .ace file, and the command name changed, and then the .ace file woudl have to be re-read into the database. This way, all we need to change is one line in the 'labace' script, to change the program.
Also note that the file to be viewed is referred to using the $ACEDB variable as a shortcut for specifying its full path. Again, if we move the database, these file paths will not need to be changed.
To view the image, click on "Pick_me_to_call". The message acedb call: $ACE_VIEW_COMMAND will appear in the window from which acedb was launched, and the image will appear on the screen.
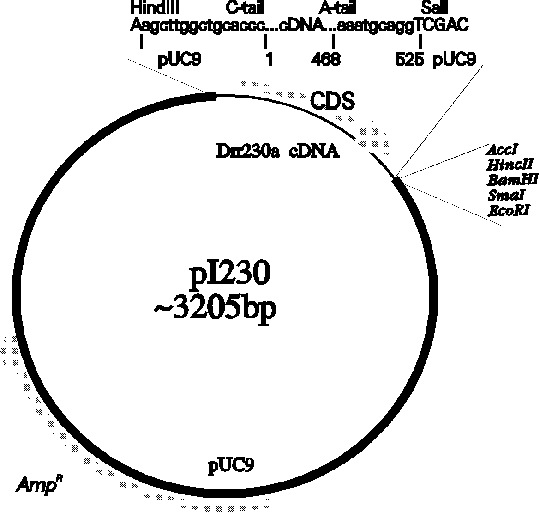
You can further familiarize yourself with ACEDB by wandering
through the different objects. This should give you a feeling for how
ACEDB works.