TUTORIAL:
Adding features to BioLegato
|
July 12, 2022 |
TUTORIAL:
Adding features to BioLegato
|
July 12, 2022 |
| cd mkdir biolegato cd biolegato |
go to your
$HOME directory create the biolegato directory go into your biolegato directory |
| gunzip
biolegato.tar.gz tar xvfp biolegato.tar |
decompresses
the file extracts the directory tree, preserving permissions and timestamps |
| directory |
purpose |
| admin |
code to set environment
variables and other settings |
| bin-linux-x86_64 | Linux - binaries placed
here will be included in $PATH |
| bin-osx-x86_64 | OSX - binaries placed here will be included in $PATH |
| blfiles | wrapper script to run
blfiles, a file manager implemented in BioLegato |
| bltable | wrapper script to run
bltable, a simple spreadsheet implemented in BioLegato |
| dat | location of PCD files used
to populate menus for each BioLegato application |
| doc | documentation |
| java | Java source code and Jar
files for BioLegato |
| local | directory for adding
BioLegato features on your local system |
| README.txt | describes contents of
current directory |
| script | executable scripts go here.
This directory is included in $PATH |
| BL_Browser='firefox' BL_PDFViewer='evince' BL_PSViewer='evince' BL_TextEditor='nedit' BL_Terminal='gnome-terminal -e' BL_ImageViewer='eog' BL_Document='oowriter' BL_Spreadsheet='oocalc' export BL_Browser BL_PDFViewer BL_PSViewer BL_TextEditor BL_Terminal BL_ImageViewer BL_Document BL_Spreadsheet |
| #------------------------
BIRCH and BIRCH_PLATFORM --------------- # # Set these two variables and everything else should work unchanged. # # In a full BIRCH system, these variables are set by getbirch as # part of an install or update. # Set BIRCH environment variable, on which everything else depends BIRCH=/home/psgendb/BIRCHDEV/java/bioLegato/test export BIRCH # Uncomment one choice for BIRCH_PLATFORM BIRCH_PLATFORM="linux-x86_64" #BIRCH_PLATFORM="osx-x86_64" export BIRCH_PLATFORM #----------------------------------------------------------------- |
 |
|
| Make sure README.txt is still selected, and
choose File --> compress. Just to
demonstrate, change Compression file format to zip. Click on
Run. |
 |
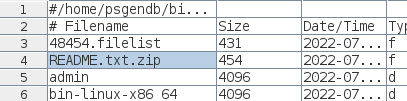
| You will see that the file has been renamed
README.txt.zip, and has shrunken to 454 bytes from the
original size of 477 bytes. (Admittedly, not much of a
change.) |
 |
| To restore the compressed file, run File
--> decompress. Note that we don't have to tell
decompress how the file was compressed. The decompression
method will be automatically determined based on the file
extension, in this case, .zip. |
 |
| The original filename is restored, and the
size is back to the original 477 bytes. |
 |
1. Select dataThe last point brings us to one of the most powerful aspects of BioLegato - the ability to send results to a new BioLegato instance, or ad hoc pipelining. To illustrate, select the script directory in blfiles, and choose File --> OpenDir - open a directory (folder).
2. Choose a program
3. Set parameters (if any)
4. Run the program
5. Work with the results.

| title=BioLegato: blfiles
- simple file manager pcd.menus.path=$BIRCH/dat/blfiles/PCD:$BIRCH/local/dat/blfiles/PCD table.generic_headers=true turtle=false |
| pcd_order |
blfiles |
| File Documentation Edit Help |
 |
| pcd_order |
blfiles |
| Refresh.blmenu OpenDir.blmenu ViewFile.blmenu Makelinks.blmenu ShortNames.blmenu ReName.blmenu GetFiles.blmenu ConcatFiles.blmenu Compress.blmenu Decompress.blmenu DeleteFiles.blmenu NewRow.blmenu SaveSel.blmenu Export.blmenu |
 |
| name
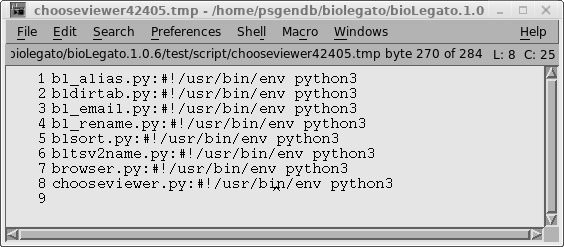
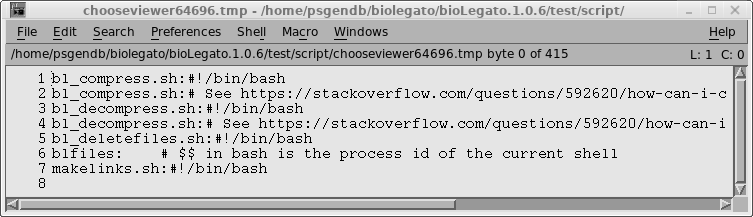
"View file" icon "$BIRCH/dat/birch_black_ico.png" tip "View a file in the appropriate program, based on the file extension." exec "bltsv2name.py %in1% %in1%.tsv; FILE=`head -n 1 %in1%.tsv`; rm %in1%.tsv%; chooseviewer.py $FILE" var "in1" type tempfile direction in format tsv save false |
| name
"Compress - compress files" icon "$BIRCH/dat/birch_black_ico.png" tip "Compress files, choosing compression method based on file extension eg. gz, zip, bz2, 7z" var "in1" type tempfile direction in format tsv save false var "ext" type chooser label "Compression file format?" default 0 choices "Gzip (gz)" "gz" "Zip (zip)" "zip" "bzip2 (bz2)" "bz2" "p7zip (7z)" "7z" var "out1" type tempfile direction out format tsv save false overwrite true content canvas panel var "Run" type button label "Run" shell "bltsv2name.py %in1% %in1%.tsv; bl_compress.sh %in1%.tsv %EXT%; bldirtab.py %out1%" close true var "Manual" type button label "Manual" shell "chooseviewer.py $BIRCH/doc/bioLegato/bl_compress.html" close false |