TUTORIAL: Genome AssemblyPreprocessing
of sequencing reads
|
November 30, 2023 |
TUTORIAL: Genome AssemblyPreprocessing
of sequencing reads
|
November 30, 2023 |
| raw read files (Illumina,
paired end) |
insert size
(nt) |
| DL300_S1_L001_R1_001_sample.fastq.gz DL300_S1_L001_R2_001_sample.fastq.gz |
300 |
| DL400_S2_L001_R1_001_sample.fastq.gz DL400_S2_L001_R2_001_sample.fastq.gz |
400 |
| DL700_S3_L001_R1_001_sample.fastq.gz DL700_S3_L001_R2_001_sample.fastq.gz |
700 |
| Launch blreads by typing 'blreads &'.
(Here, we're adding & to the blreads command to run blreads in the background. That will allow us to continue using the command line in the same terminal window in which blreads was launched.) Note that the path for the current working directory is listed on the first line of blreads. Any line begining with a hash mark (#) is a comment, and will be ignored. |
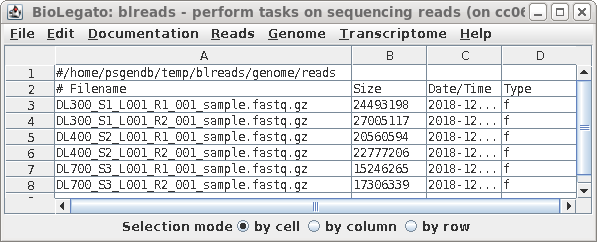 |
| Choose the first file to which you want to
make a link. To make the name of the link shorter, type in a target pattern that is common in two or more files. At right, the pattern is "_S1_L001". Since the short pattern field is left blank, this string will simply be omitted from the link name. The link name will be DL300_R1_001_sample.fastq.gz. |
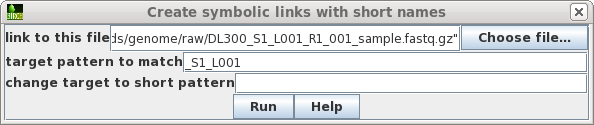 |
| The name of the link will appear in the
blreads window. Note that the type field of the original
file has 'f', to indicate a file, and the type field for the
link is 'l'. to indicate a link. |
| When you have completed the process for all
six files, blreads should look like this: For each set of paired-end read files (R1 and R2), there will be a corresponding pair of symbolic links with shorter names. |
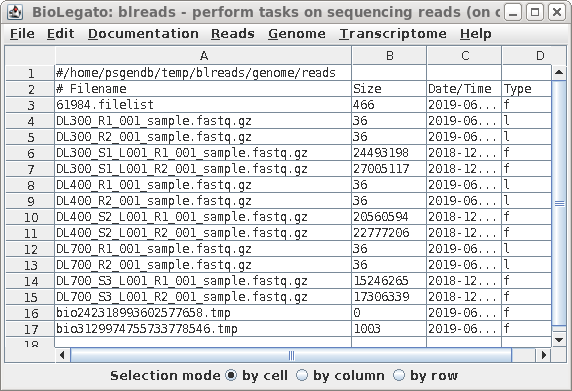 |
| To avoid accidentally changing the names of
the original files, first sort the files based on the Type
field, so that all links appear together in blreads. Choose
Edit --> BLSORT. Set the 1st sort key to column 4
(ie. Type), and Sort order to Ascending. Choose Run:Output
to this window. |
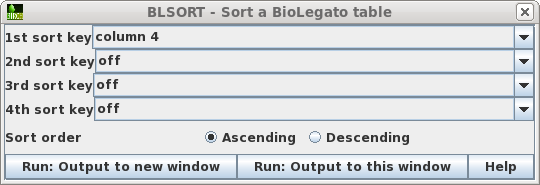 |
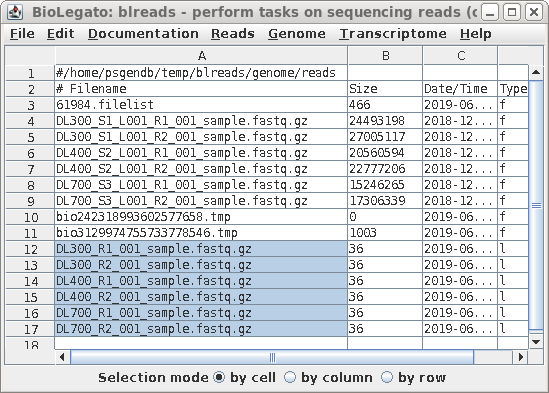
| All links will now be together in blreads.
Select the links as shown, and choose File --> Rename.
|
 |
| Set the target pattern to '_001_sample', and press Run. | 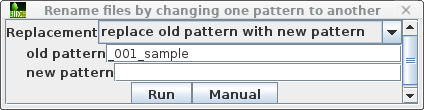 |
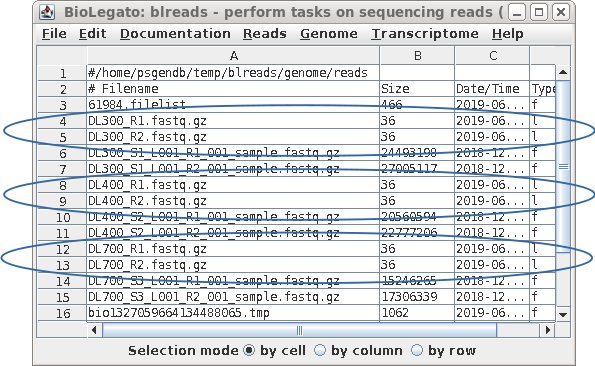
| Each pair of read files now has short, easy
to distinguish names, that will be used in all subsequent
steps. |
 |
| By default, the number of threads used is 1/4
of the number of available cores, or 1, whichever is
greater. For most datasets, this is not a very time
consuming step. Therefore, it is usually unnecessary to use
additional cores. Simply click on Run. |
 |
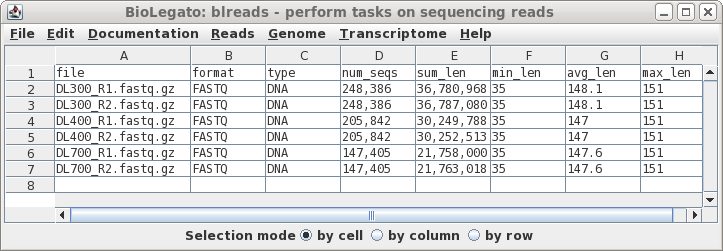
| In this case, all of the files look fairly
similar in terms of the number of reads and the average
length of reads. At this point, if there was a file that looked to be aberrant and you knew that it should be discarded, simply select the name of the bad file and delete the link using File --> Deletefiles. |
 |
| Typical read files are larger than this
sample dataset, so with larger files, you may wish to speed
up FASTQC by setting a larger number of cores. In most
cases, results will appear soon enough that you don't need
to set notification of completion by email. Simply click on Run. |
 |
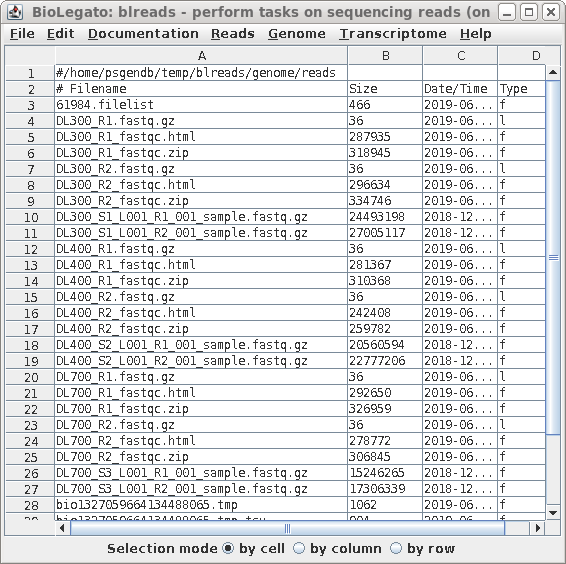
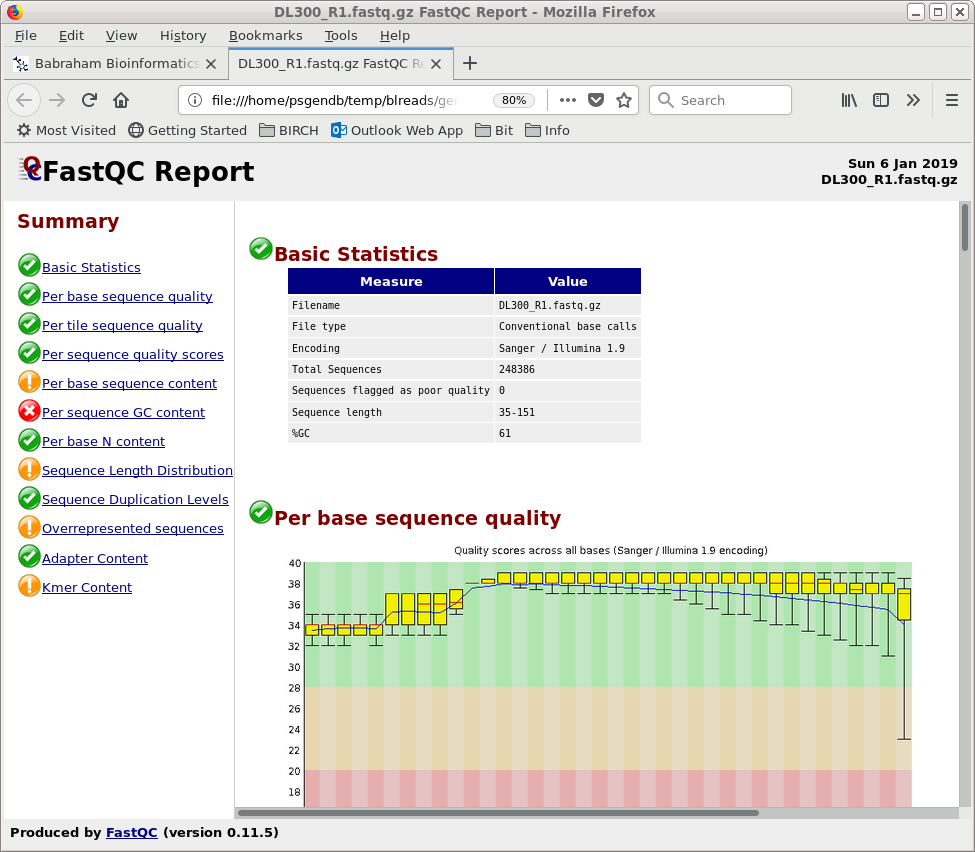
| blreads now lists the output files from
FASTQC. Files with the .html extension are viewable in any
web browser. The .zip files are the accompanying graphics
used by the web files. To view any report, select an HTML file and choose File --> View file. The HTML file will pop up in the browser. |
 |
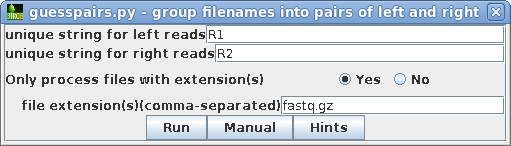
| Most Illumina services use R1 and R2 as the
substrings which distinguish the left and right read pair
files for a given library, so by default, R1 and R2 is set
for "unique string for left/right reads" We only want to process read files, and not the other files in the directory. For this dataset, all read files use the fastq.gz extension. Click on Yes and type in fastq.gz. Clicking on the Hints button will give a more detailed explantion of these parameters. |
 |
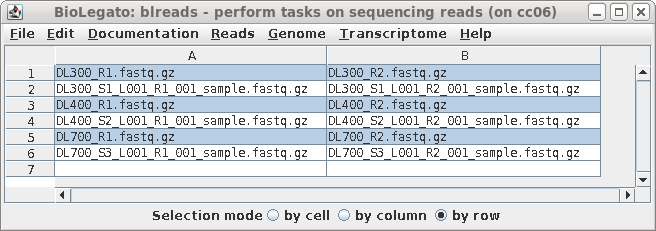
| A new blreads window will appear with the
left and right read files in two columns. Both the symbolic
links and the long-named read files appear. Since we prefer
to work with the short-named links, these can be selected by
clicking on Selection mode: row below, and then clicking on
the short-named read files. (This accomplishes the same
thing as sorting did above.) |
 |
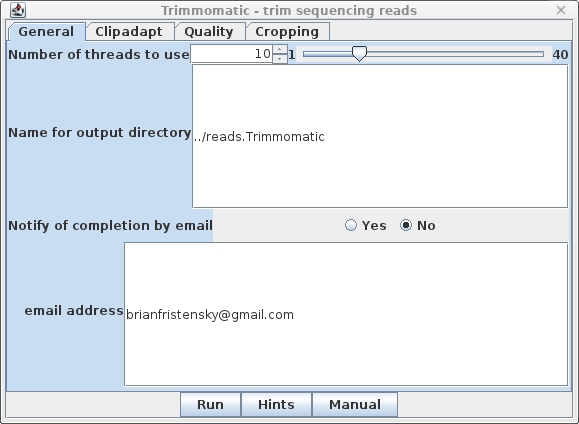
| General - The parameters for
Trimmomatic are grouped into four tabs. The General tab has
basic settings. By default, output will go to the
../reads.Trimmomatic directory. Trimmomatic performs each of the processing steps in an order specified by the user. Individual parameters can be turned on or off, and the order in which they are performed is set by the rank parameter. |
 |
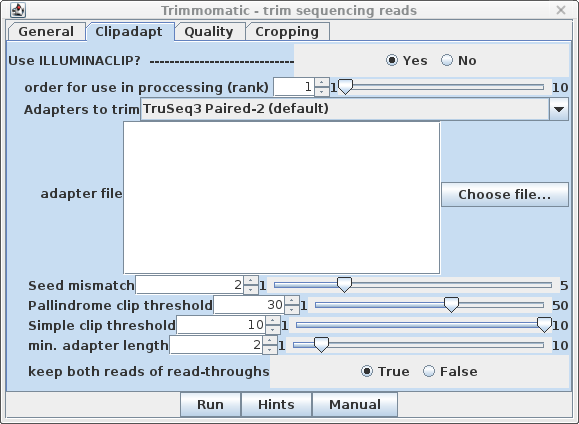
| Clipadapt - By default, the
ILLUMINACLIP step is performed first (ie. rank=1). The
defaults are those given in the Trimmomatic manual, with the
exception that keep both reads of read-throughs is
set to true. This may help to avoid cases where a single
read of a read pair is deleted during trimming, which can
cause some transcriptome assembly programs to crash. |
 |
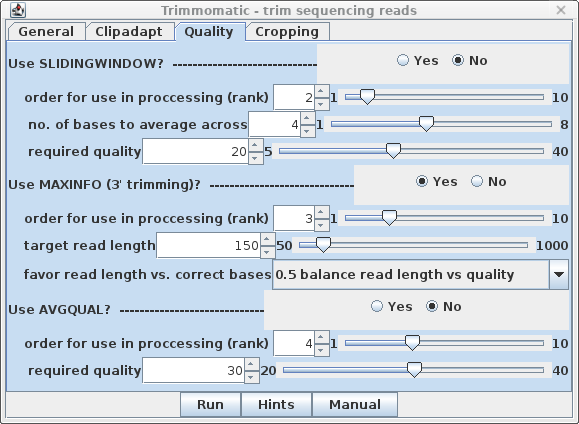
| Quality - By default, SLIDINGWINDOW
and AVGQUAL are off. Set MAXINFO to Yes. Since rank
is set to 3, we don't need to change this for MAXINFO to be
performed as the next step. |
 |
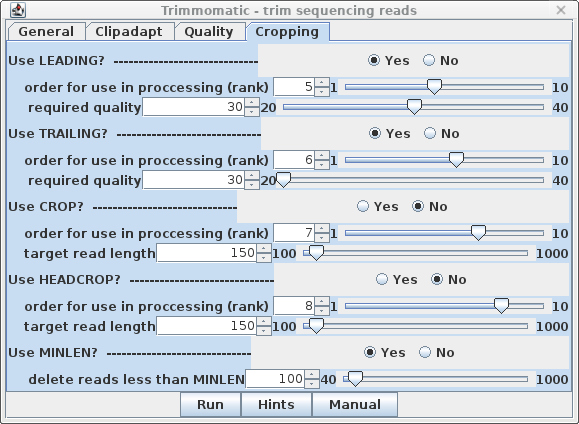
| Cropping - It's a good idea to
eliminate poor quality nucleotides from 3' and 5' ends of
reads, so set LEADING and TRAILING to Yes.
Set the required quality score to trim at 30, rather than
20, which will shorten some reads, but the remaining bases
will be of higher quality. Finally, the last step is MINLEN, which eliminates read pairs below a specific length. For 150 bp reads a value of 100 is a reasonable compromise between very short reads, which would be difficult or impossible to uniquely map to transcripts, and much longer reads, which might compromise coverage of reads located in the 5' ends of transcripts. No rank is assigned, because this step must be done after all other trimming steps are completed. |
 |
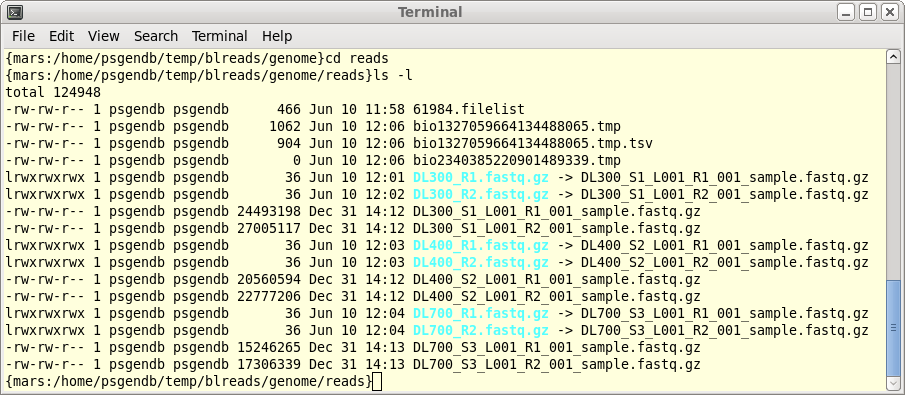
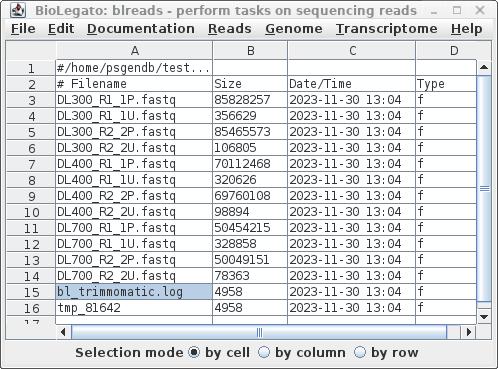
| When Trimmomatic has processed all files, a
new blreads window will pop up. It is important to note that
this new window is running in the reads.Trimmomatic
directory. At this point, one can probably close the
previous blreads window, which is running in the reads
directory. The next steps will be done in the
reads.Trimmomatic directory. First, notice that by default, trim_galore automatically runs FASTQC, so we have .zip and .html reports for each set of reads after trimming. For each of the original read files, there are now two output files.
|
 |