October
31, November 5, 2024
DATABASES
REFERENCES
Harrington, JL (2000) Object-Oriented
Database
Design Clearly Explained. Morgan Kaufmann Publishers.
Simon, AR (1995) Strategic
Database
Technology. Morgan Kaufmann Publishers.
Walsh S and Anderson M
(1998) ACEDB: A database for genome information. In
Baxevanis AD and Ouellette BFF. Eds. Bioinformatics: A
practical guide to the analysis of genes and proteins .
John Wiley, Toronto.
A.
WHY DATABASES?
B.
TYPES OF DATABASES
1. Flatfile
2. Relational Databases
3. Object-oriented databases
4. Hypertext databases
C.
ACeDB
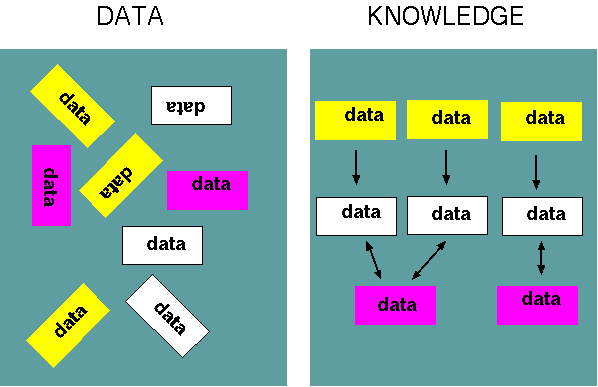
Energy can be a metaphore for data:
"Among the unconventional sources of energy,
conservation presents itself as the most immediate
opportunity. It should be regarded as a largely untapped
source of energy. Indeed, conservation - not coal or nuclear
energy - is the major alternative to imported oil. "
Stoboagh, R
and Yergin D (1979) Energy Future. pg. 10.Ballantine
Books, New York.
A.
WHY DATABASES?
- To
collect and preserve data
- To make
data easy to find and search
- To
standardize data representation
- To make
data machine-readable
- To
organize data into knowledge
B.
TYPES
OF DATABASES
1.
Flatfile databases
A flatfile database
consists of a collection of records, each containing several
data fields.
For example, three
records in a database of cell stocks might look like this:
Cell_Stock : "SK11.pEA215.3"
Species "Escherichia coli"
Plasmid "pEA215.3"
Experiment "SK11"
Freezer "AG334 -80C"
Box "Pisum ESTs II"
Gridded "Rack(BF7) Box(Pisum ESTs II)"
Cell_Stock : "SK11.pI206KS"
Species "Escherichia coli"
Plasmid "pI206KS"
Experiment "SK11"
Freezer "AG334 -80C"
Box "Pisum ESTs II"
Gridded "Rack(BF7) Box(Pisum ESTs II)"
Cell_Stock : "SK11.pEA46.2"
Species "Escherichia coli"
Plasmid "pEA46.2"
Experiment "SK11"
Freezer "AG334 -80C"
Box "Pisum ESTs II"
Gridded "Rack(BF7) Box(Pisum ESTs II)"
One observation that can
be made immediately is that flatfile databases are highly redundant. In
this example, the data in the Species, Freezer, Box and Gridded
fields are identical. Not only does this redundancy waste space,
it means that if a field has to change, it must be changed in all
records. For example, if the box was moved to a different freezer,
all Freezer fields for that box would have to be changed.
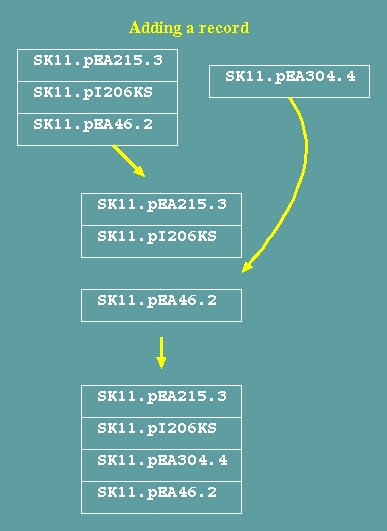
Transactions update the database
Transactions that
change, add or delet records require rewriting the database. To
add a record, the database is written to the point at which the
record is to be added, the new record is written, and then the
rest of the records is written. Similarly, changing and deleting
records also requires that all records be rewritten.
Searching the database is done by linear search or index search.
Searching a flatfile
database normally requires that the entire text of the database be
searched. Faster searches can be done if the database is indexed.
For example, the Cell Stock database above could be indexed by
Experiment, Plasmid, Species, etc. Indexing can greatly
increase the size of the database.
Flatfile databases force a single view of the data
Probably the single most
important disadvantage of flatfile databases is that the records
must be organized by one of the fields. In the example, the
decision would have to be made that the Cell Stock was the
fundamental unit of data, and that other types of information,
such as Plasmids or Species, were best considered as an attribute
of a Cell Stock. For many purposes, this is an unnatural
representation of the data.
GenBank entries
retrieved by Entrez are flatfile representations of data that
are centered around DNA sequences. In a GenBank entry, every
field appears to belong to a particular sequence. We will see
later that the flat file representation is simply one possible
view of a richer database.
2.
Relational databases
Relational databases can
be thought of as comprehensive tables of data. Every record from a
flatfile could be implemented as a row in a table. The Cell Stock
database might be implemented as a table with
Cell_Stock Experiment plasmid Freezer Box Species
SK4.pcyclinB SK4 pcyclinB AG334 -80C Pisum ESTs Escherichia coli
SK4.pcyclinD SK4 pcyclinD AG334 -80C Pisum ESTs IEscherichia coli
SK4.pdiminuto SK4 pdiminuto AG334 -80C Pisum ESTs IEscherichia coli
SK4.pFF100 SK4 pFF100 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pME1 SK4 pME1 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pNDK-P1 SK4 pNDK-P1 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pPABP1 SK4 pPABP1 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pPCNA SK4 pPCNA AG334 -80C Pisum ESTs IEscherichia coli
SK4.pPS-IAA4-5 SK4 pPS-IAA4-5 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pPS-IAA6 SK4 pPS-IAA6 AG334 -80C Pisum ESTs IEscherichia coli
SK4.pTic110 SK4 pTic110 AG334 -80C Pisum ESTs IEscherichia coli
From a relational
database, many views of the data are possible.
Plasmid View
Plasmid Species Cell Stock
pEA25 Escherichia coli SK10.2.pEA25
pEA46.2 Escherichia coli SK11.pEA46.2
pEA207.2 Escherichia coli SK11.pEA207.2
pEA214.6 Escherichia coli MB123.pEA214.6
pEA215.3 Escherichia coli SK11.pEA215.3
pEA238.2 Escherichia coli MB123.3.PEA238.2
pEA238.11 Escherichia coli MB123.3.pEA238.11
pEA277.11 Escherichia coli SK11.pEA277.11
pEA303.4 Escherichia coli SK11.pEA303.4
pEA315.2 Escherichia coli MB123.3.pEA315.2
peB4 Escherichia coli VB1.eB4
Experiment View
Experiment Cell
Stock
Box
Freezer
SK4
SK4.pPS-IAA4-5
Pisum ESTs I AG334 -80C
SK4
SK4.pPS-IAA6
Pisum ESTs I AG334 -80C
SK4
SK4.pTic110
Pisum ESTs I AG334 -80C
SK4
SK4.pToc34
Pisum ESTs I AG334 -80C
SK4
SK4.pToc86
Pisum ESTs I AG334 -80C
SK5
SK5.pAB96.3
Pisum ESTs I AG334 -80C
SK5
SK5.pABR17.10
Pisum ESTs I AG334 -80C
SK5
SK5.pABR18.2
Pisum ESTs I AG334 -80C
SK5
SK5.pI39
Pisum ESTs I AG334 -80C
SK5
SK5.pI49KS
Pisum ESTs I AG334 -80C
SK5
SK5.pI176KS
Pisum ESTs I AG334 -80C
SK5
SK5.pI225KS
Pisum ESTs I AG334 -80C
Relational
databases
can be organized across many files
Although a relational
database can be implemented in a single large table or "relation",
it is often advantageous to split the database up into multiple
tables.
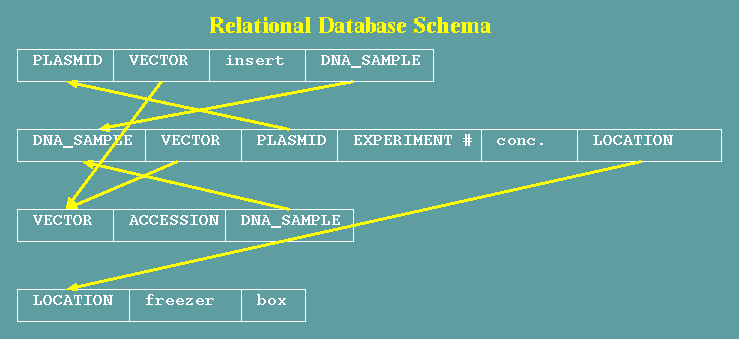
Four tables are shown:
PLASMID, DNA_SAMPLE, VECTOR and LOCATION. Fields in lowercase
contain text or other data. Fields in uppercase contain names of
items in other tables. Fields that reference other tables are
referred to as links. Several factors have to be considered when
designing a relational database:
- Text fields -
Text fields contain data. For example, in the location
table, 'freezer' might contain the name of a freezer, and box
would contain whatever was written on the box containing the
sample.
- Reference
fields - Reference fields contain the name of records
in another table. The PLASMID field in DNA_SAMPLE tells which
plasmid the sample came from. Conversely, the DNA_SAMPLE field
in the PLASMID table tells which
- Links have
directions - In the example, each link can be made in
either one or both directions. DNA_SAMPLE has a LOCATION field
that points to a LOCATION, but LOCATION does not point back to
any given DNA_SAMPLE. This link is therefore a 1-way link. In
comparison, The relationship between PLASMID and DNA_SAMPLE is
two-way.
- One-to-one vs.
one-to-many - If two-way links are permitted, the
possiblity exists for one-to-many relations. For example,
there may be many DNA_SAMPLES for a VECTOR or PLASMID.
Implementation for one-to-many relations requires that the
underlying database software contains methods for creating
one-to-many links.
- Choice of data
types - In this database, the distinction is made
between plasmid and vector. While some plasmids may be
considered vectors, not all vectors are plasmids (eg. Lambda
phage). There is the further distinction between a vector,
which is used for cloning, and a plasmid construct, which is a
vector plus an insert.
- Alternative
links - The DNA_SAMPLE table has both VECTOR and PLASMID
fields. In the current implementation of the database, these
are not mutually exclusive. The structure of the database
permits a DNA_SAMPLE to come from a PLASMID and a VECTOR at
the same time. From the researcher's viewpoint, this probably
makes no sense, and in practice only one of the fields would
be used in any given DNA_SAMPLE. Ideally, the database
software should have a way to create fields that have
alternative, mutually-exclusive forms.
- Independence of
tables - Having separate tables has advantages and
disadvantages. Modification of text fields in one table has no
effect on other fields. In fact, any number of text fields or
reference fields can be added to one table without
invalidating other tables. For example, adding text
information to PLASMID such as size of the plasmid, or
insertion site for the insert, make no difference to the
DNA_SAMPLE definition. However if we decide that a DNA_SAMPLE
could also be an oligonucleotide, we could create a new table
called OLIGO. In this case, both DNA_SAMPLE and OLIGO would
have to have fields that point to each other.
- Modifications
are done on specific tables - One advantage of
relational databases is that by breaking up the database into
many tables, in many cases only one table needs to be
rewritten when making changes in fields. In other cases,
addition of a record may require rewriting many or most
tables.
Common relational database packages:



 Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada
Unless otherwise cited or
referenced, all content on this page is licensed under
the Creative Commons License Attribution
Share-Alike 2.5 Canada