What Mesquite Does
Mesquite is software for evolutionary biology, designed to help biologists manage and analyze comparative data about organisms. Its emphasis is on phylogenetic analysis, but some of its modules concern population genetics, while others do non-phylogenetic multivariate analysis. Because it is modular, the analyses and management features available depend on the modules installed. Here is a brief overview of some of Mesquite's features. See also a more complete outline of features, and the Mesquite Project Youtube channel, with instructional videos helping you learn Mesquite.
Despite Mesquite's broad analytical capabilities, the developers of Mesquite find that we use Mesquite most often to provide a workflow of data editing, management, and processing. We will therefore begin there.
Data Management and Processing
Mesquite has many features for managing data. It has an editor for editing morphological data: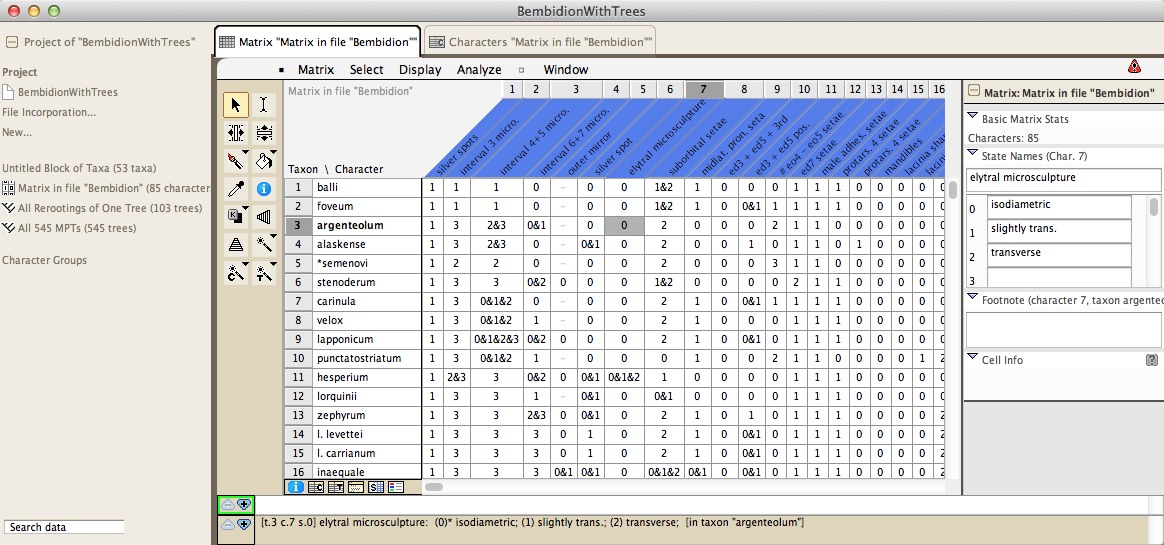
and DNA and protein sequence data:
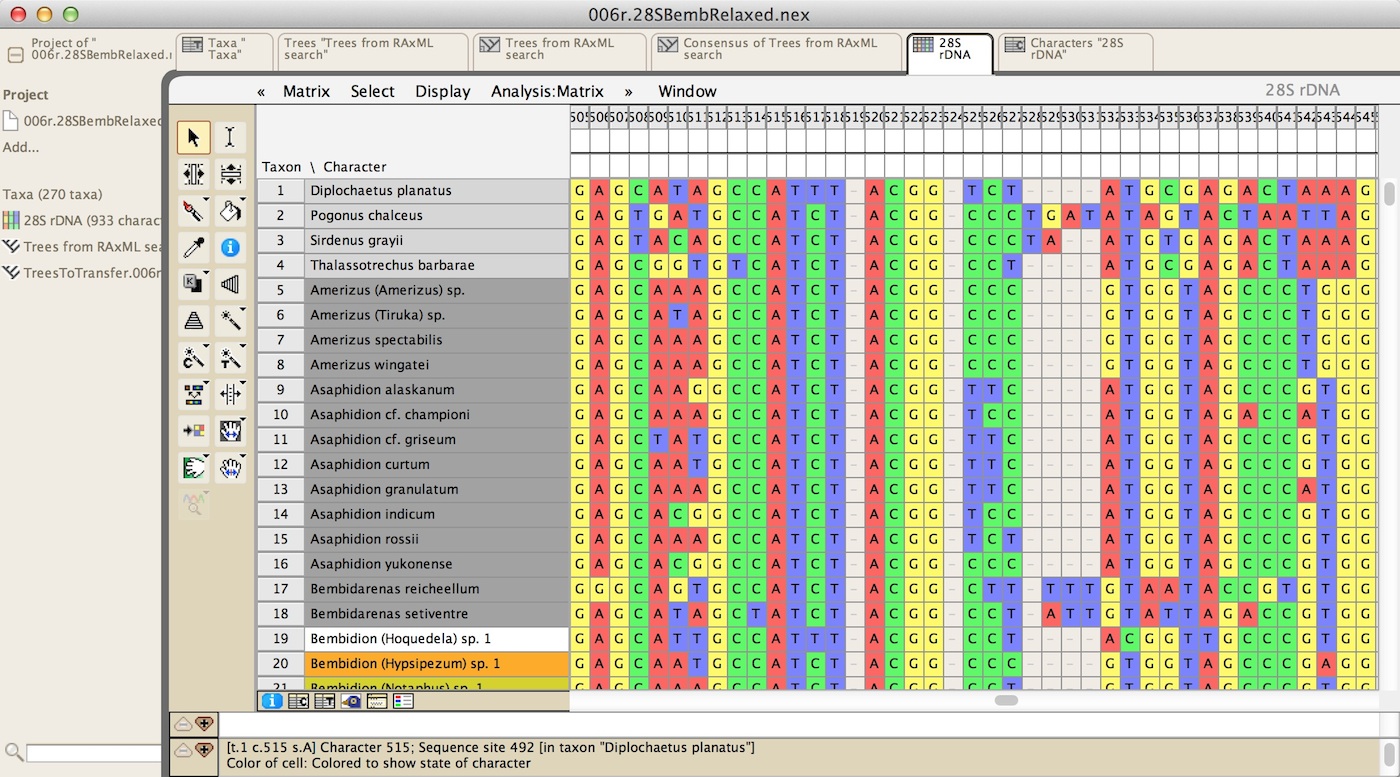
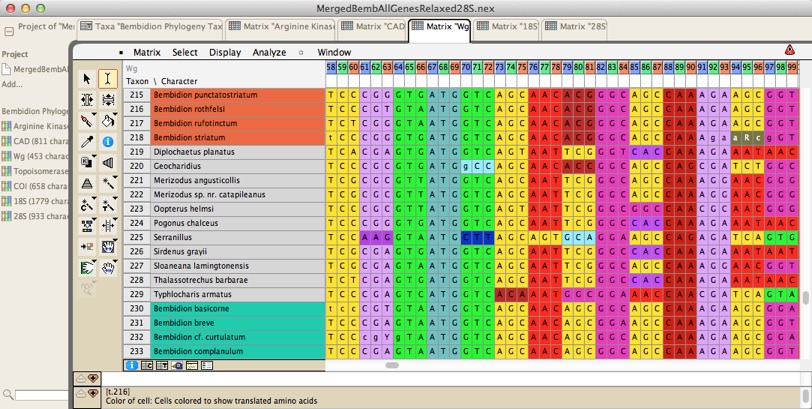
There are tools for manual and automated sequence alignment, as well as many other tools to manipulate the sequences. There are also many visualization options; for example, here are some protein-coding DNA sequence with the colors of the cells representing the amino acids for which the nucleotides code:
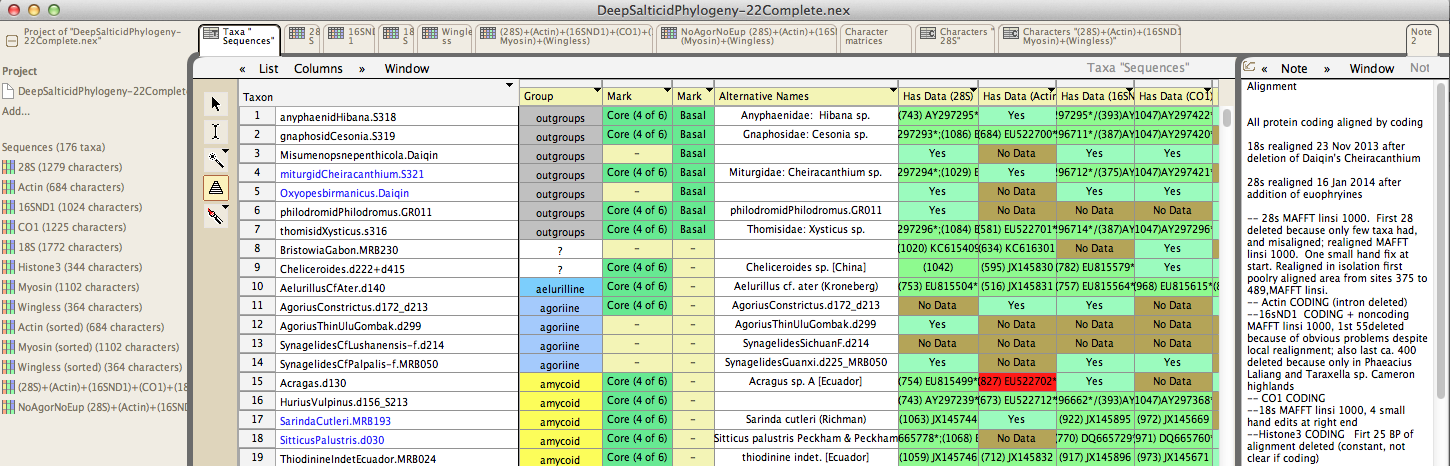
Metadata of many sorts can be attached to taxa, characters or cells in the matrix, including annotations, images, alternative names, pre-selected sets, and colored labels. All of these help with sorting and manipulating data:
Trees & Analyses
Mesquite has many features for visualizing and editing trees. It is not primarily designed to infer phylogenetic trees, but rather for diverse analyses using already inferred trees.Mesquite has a tree viewer and editor, from which you can print publication-quality trees:
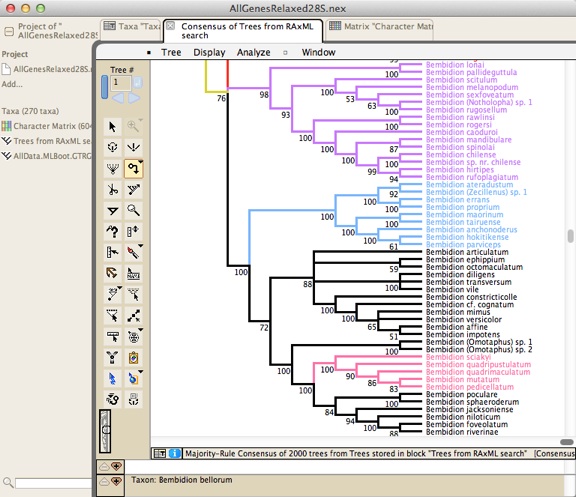
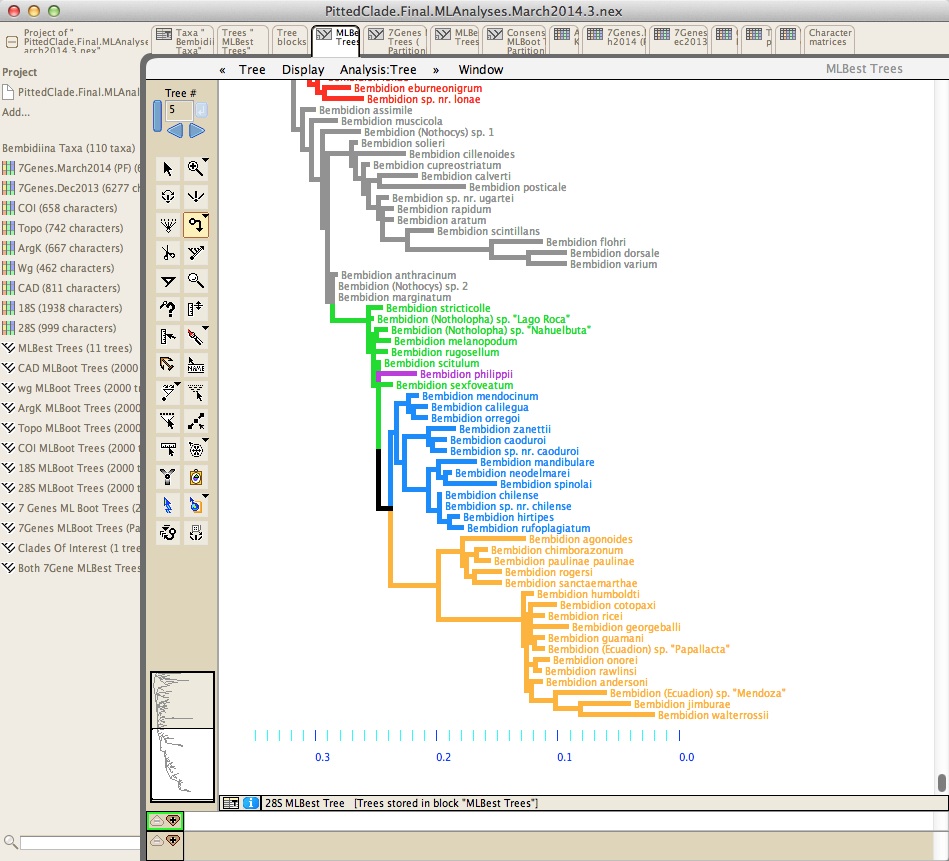
Analyses using phylogenetic trees can be done in the tree window, such as reconstruction of ancestral states:
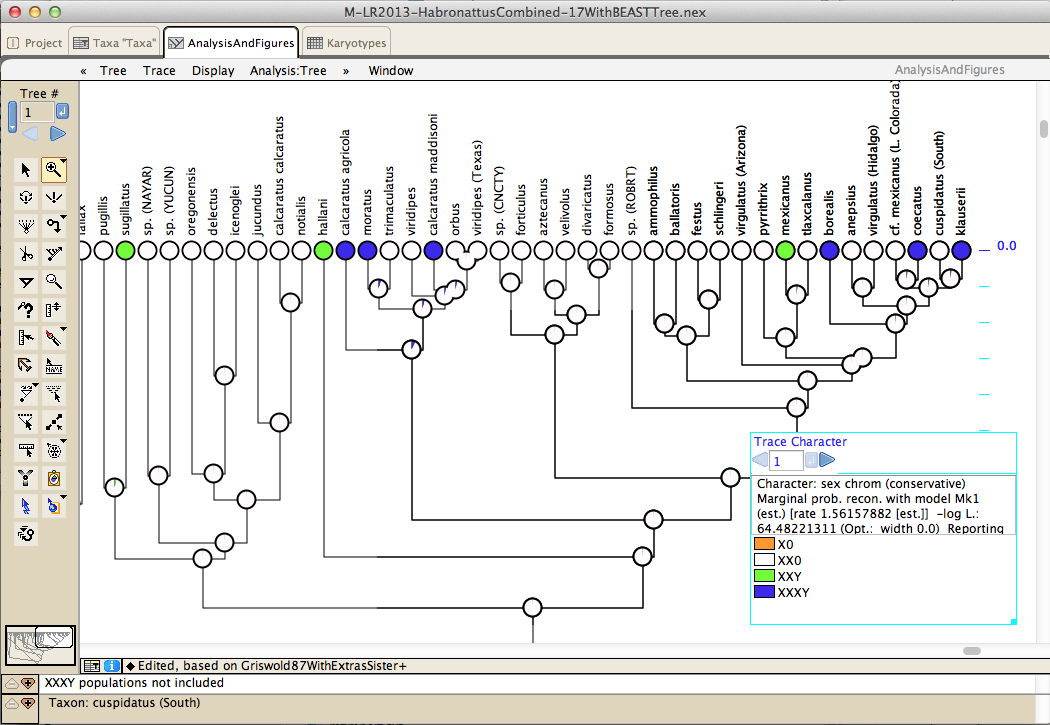
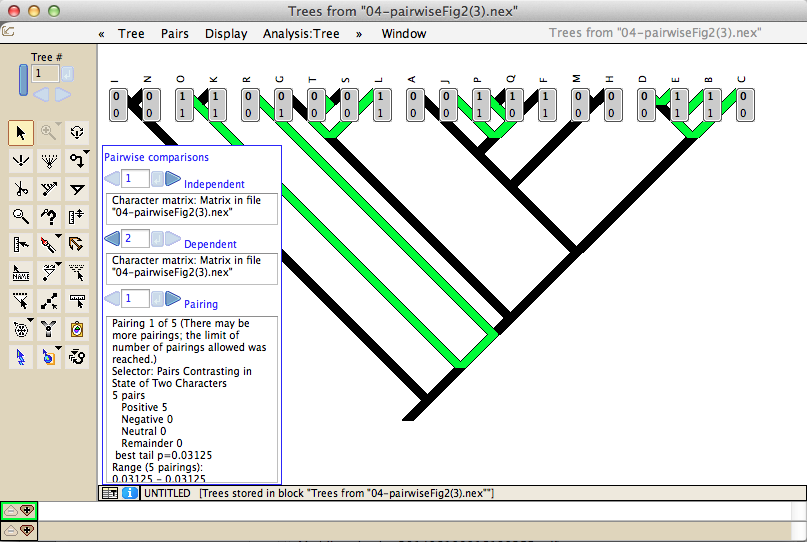
Analyses to test processes of character evolution include Pagel's 1994 correlation test and pairwise comparisons:
Tests of species diversification like BiSSE are available:
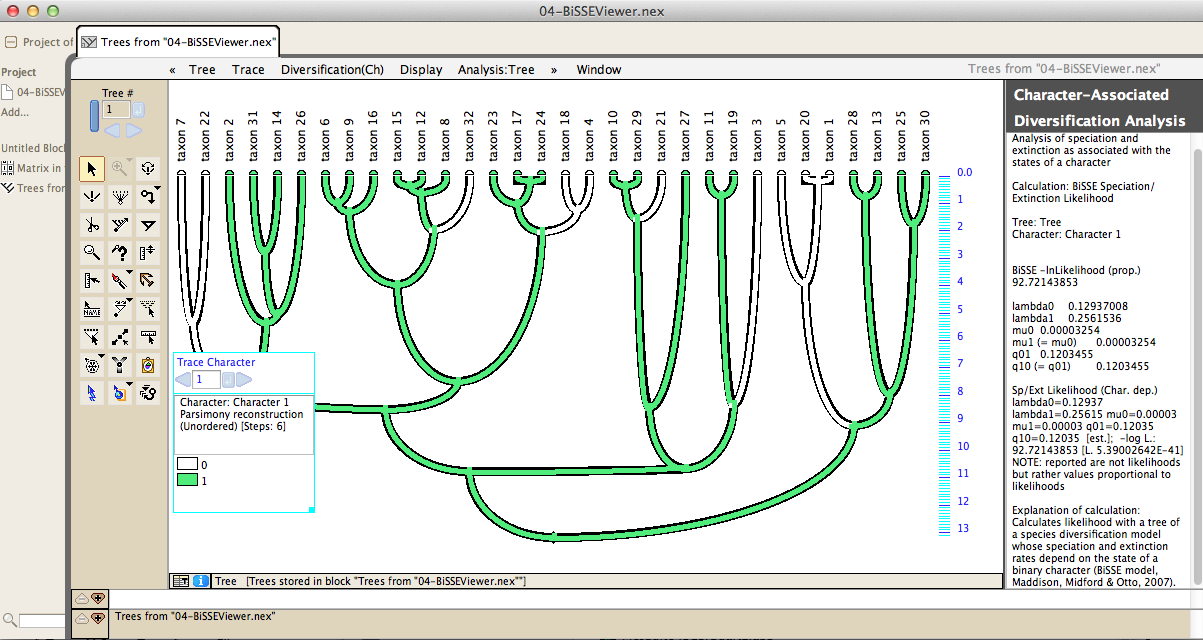
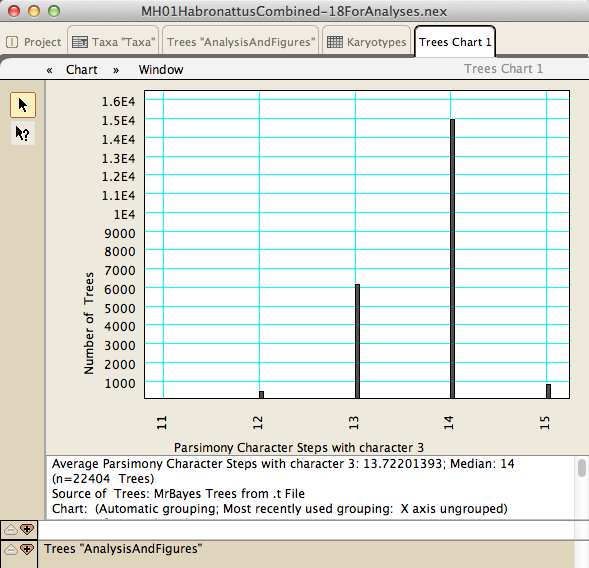
Using Mesquite's charting feature, statistics can be calculated over a suite of many trees, permitting one to determine the effect of phylogenetic uncertainty on conclusions:
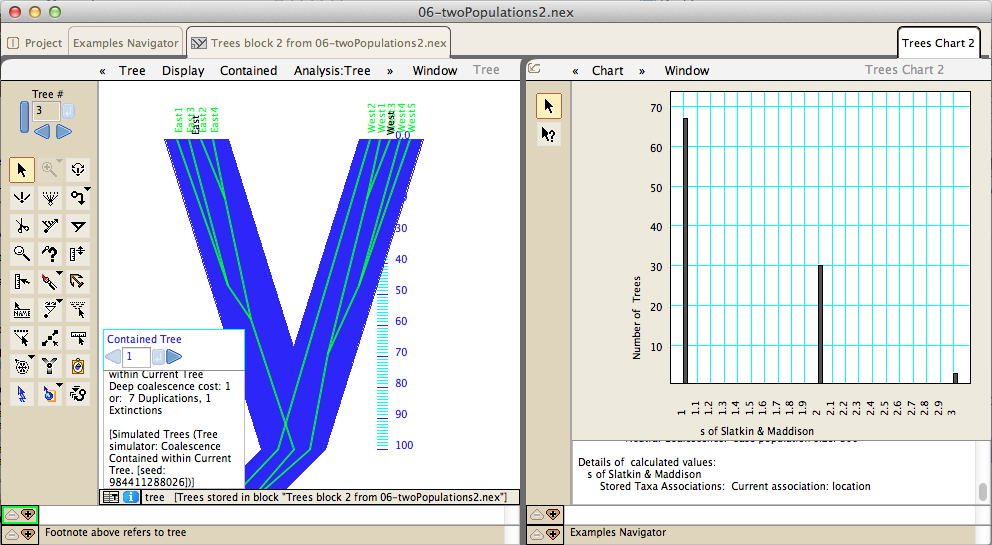
Coalescence simulations permit tests of population genetics and phylogeographic hypotheses:
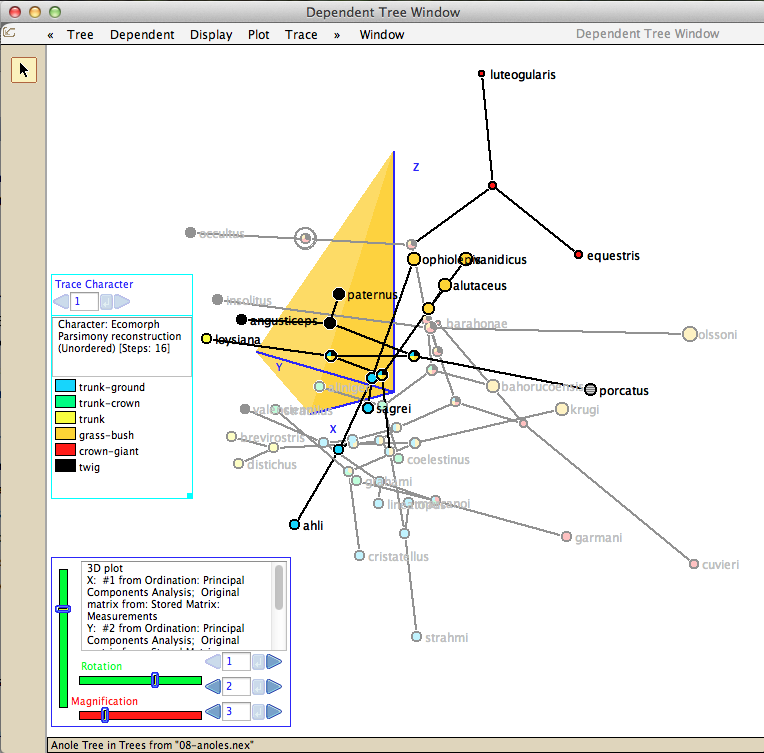
Multivariate analyses including PCA and CVA can be done, and the phylogenetic tree embedded in morphospace:
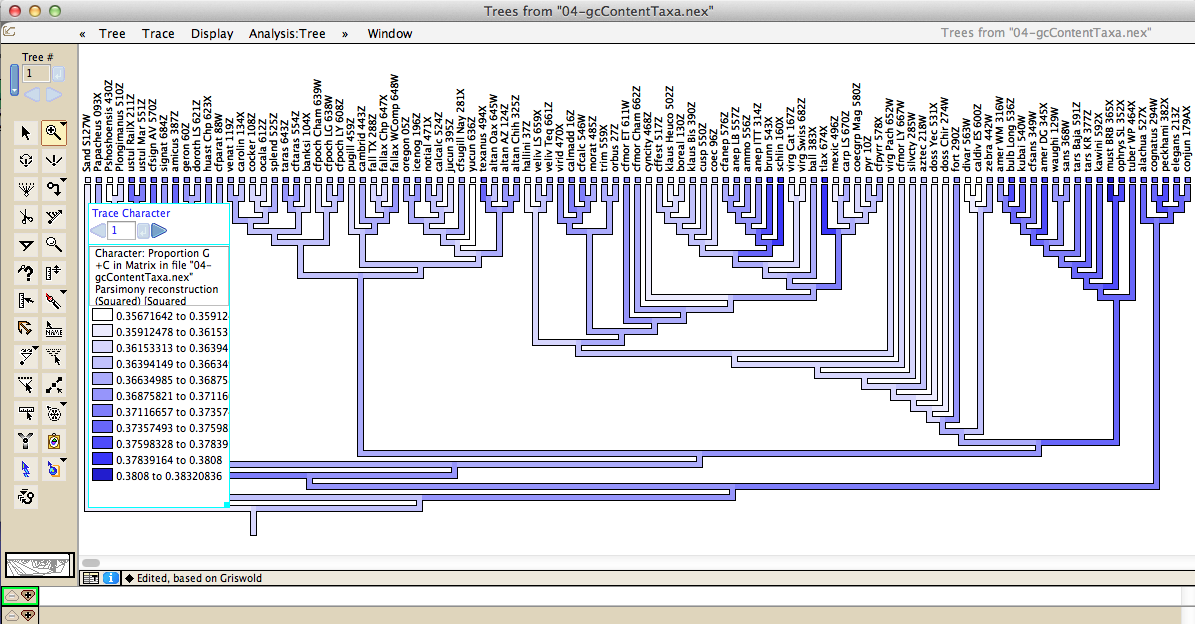
Various other statistics and be visualized on trees, such as GC/AT compositional bias:
There are other analyses as well, including:
- Simulation of character evolution (categorical, DNA, continuous)
- Parametric bootstrapping (integration with programs such as PAUP* and NONA)
- Tree comparisons and simulations (tree similarity, Markov speciation models)
Extending Mesquite
Mesquite's modularity means that it can be extended by adding modules available in separately distributed packages. For example, there is a package called Chromaseq that provides tools for processing chromatograms and making base calls: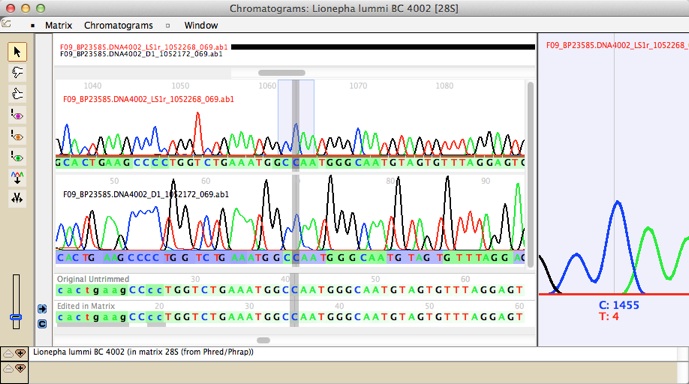
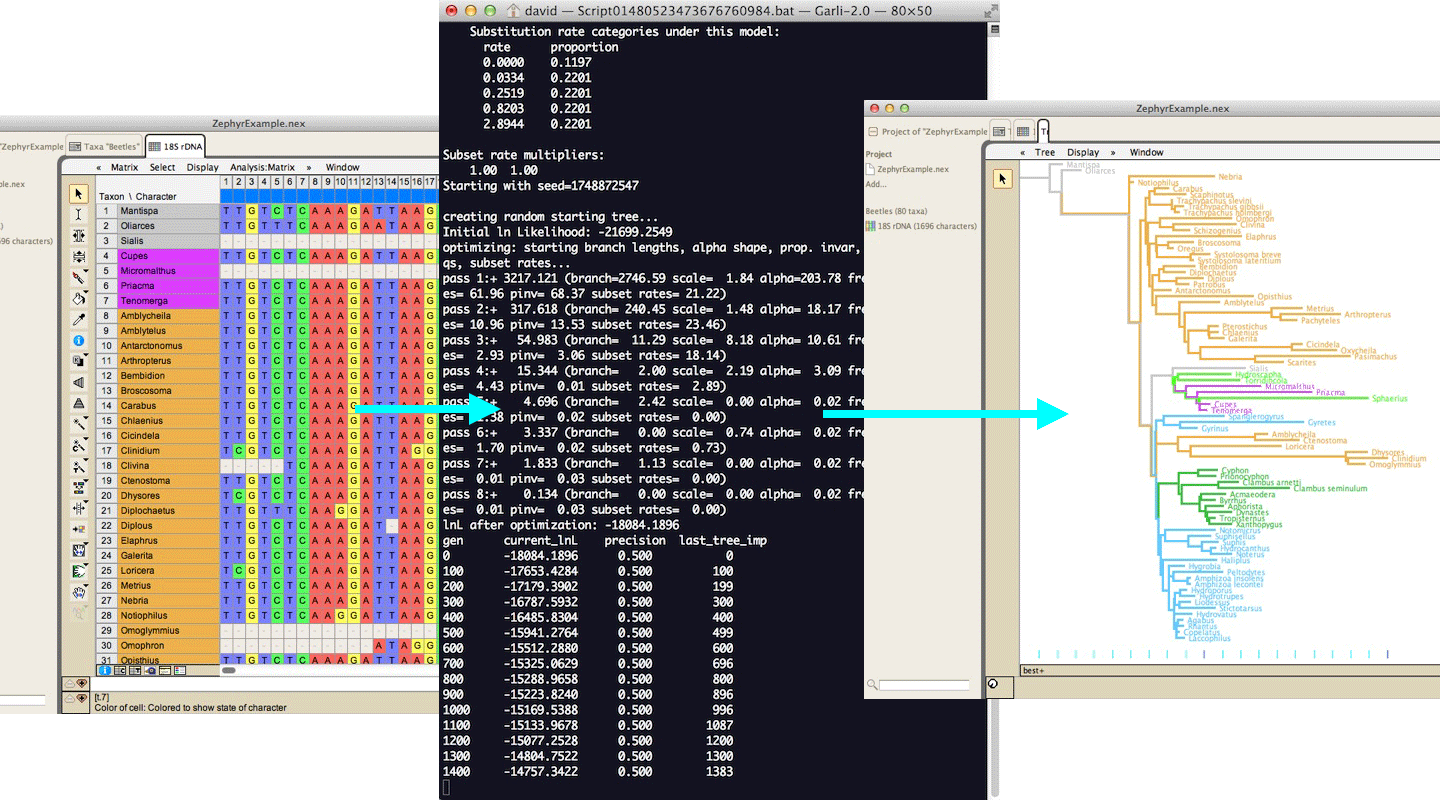
Zephyr allows Mesquite to interact with external phylogeny inference programs such as RAxML or GARLI; Zephyr sends data to these programs and then harvests the results. For GARLI, trees can be visualized as the search progresses:
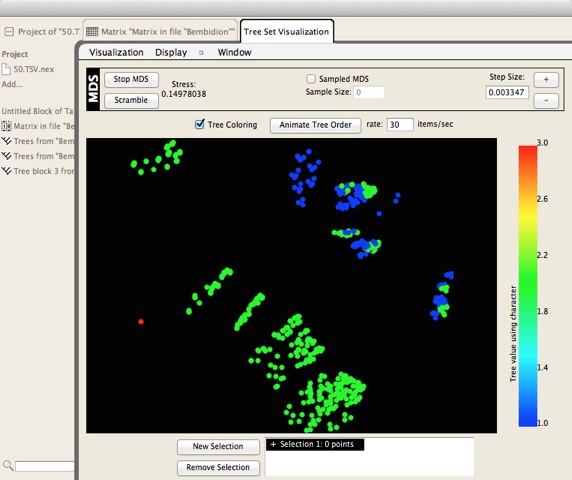
There is another package called TreeSetViz that allows you to visualize the relationships between trees in a multi-dimensional space of tree similarities:
Cartographer provides tools for mapping of taxa:
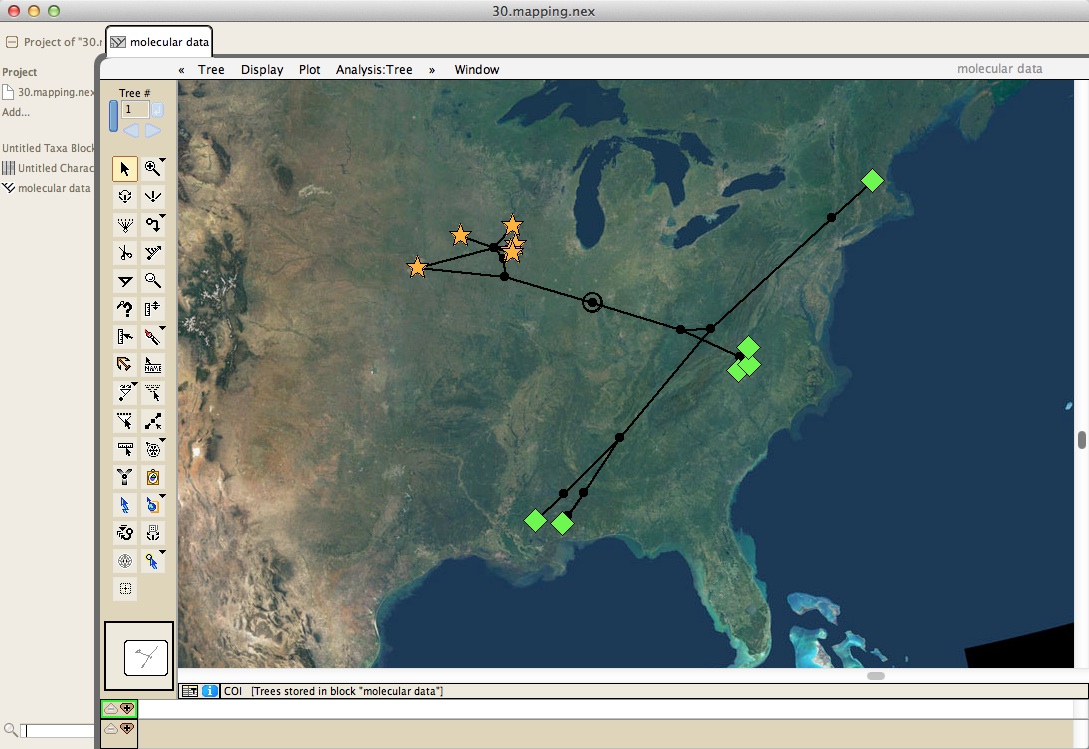
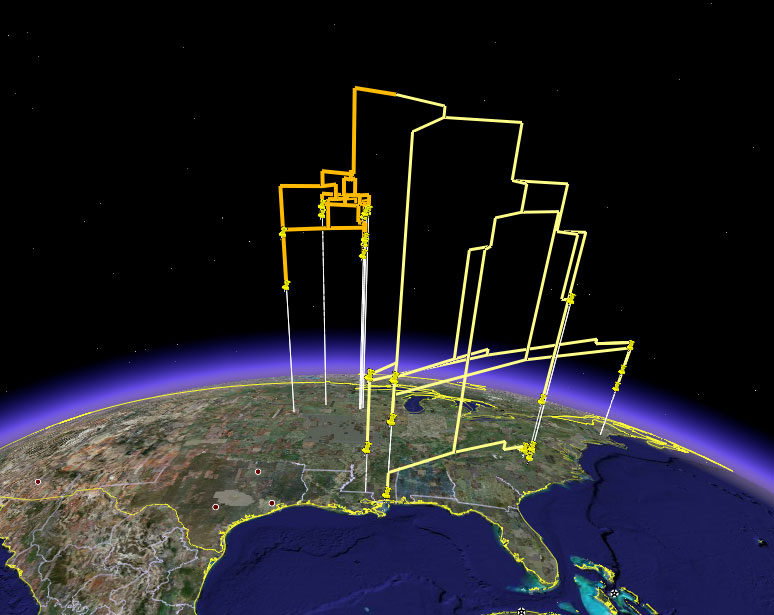
including the production of KML files that allow you to view a tree in Google Earth:
Some of the additional packages available for Mesquite are listed here.