BioLegato:
|
Oct. 21, 2014 |
BioLegato calls
programs
that run as external jobs. In essence, any program
can be run from BioLegato. Programs can run on a local
server, or as a
remote
web service. Output is returned to the user's desktop in the
appropriate viewer.
(For more on the nuts and bolts of BioLegato, go the the BioLegato Project.)
(For more on the nuts and bolts of BioLegato, go the the BioLegato Project.)

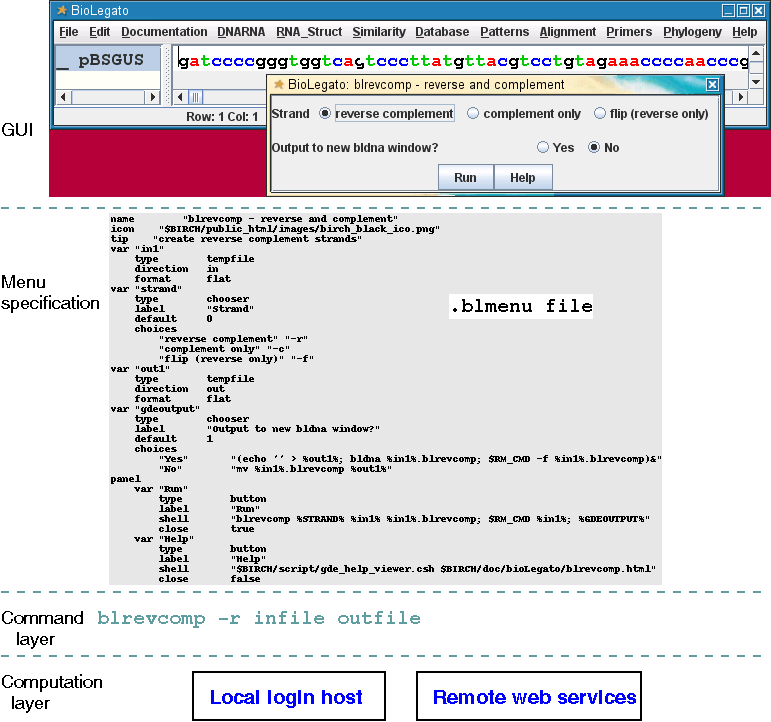
The GUI layer - BioLegato is the GUI layer. BioLegato is nothing more than a program that creates menus and calls other programs. Data selected in the BioLegato window is written to a temporary file. A Unix command is generated by BioLegato that calls a program, using the temporary file as input and supplying parameters as set in the menu. Output from the program is imported back into BioLegato, or sent to other programs for viewing by the user.
The Menu specification layer - When BioLegato starts, it reads a set of files which specify how to construct the menus that run programs. Menus are written in an easy to use language called PCD. For example, in BioLegato, the menu for DNARNA --> blrevcomp - reverse and complement menu looks like this:
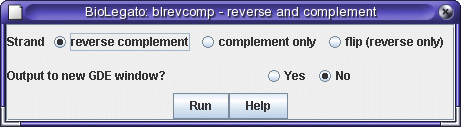
The corresponding entry in the .BLrevcomp.blmenu file creates this menu:
name "blrevcomp - reverse and complement"This menu exports the sequence to a temporary file, and uses that file as input for the blrevcomp program. The Unix command to accomplish this task is on the shell line. The command is built by substituting values from the other fields (eg. %in1%, %STRAND%, %GDEOUTPUT%) into the itemmethod line. After removing some temporary files, the commands specified in GDEOUTPUT send the output from blrevcomp either to a new BioLegato window, or to the current BioLegato window..
icon "$BIRCH/public_html/images/birch_black_ico.png"
tip "create reverse complement strands"
var "in1"
type tempfile
direction in
format flat
var "strand"
type chooser
label "Strand"
default 0
choices
"reverse complement" "-r"
"complement only" "-c"
"flip (reverse only)" "-f"
var "out1"
type tempfile
direction out
format flat
var "gdeoutput"
type chooser
label "Output to new bldna window?"
default 1
choices
"Yes" "(echo '' > %out1%; bldna %in1%.blrevcomp; $RM_CMD -f %in1%.blrevcomp)&"
"No" "mv %in1%.blrevcomp %out1%"
panel
var "Run"
type button
label "Run"
shell "blrevcomp %STRAND% %in1% %in1%.blrevcomp; $RM_CMD %in1%; %GDEOUTPUT%"
close true
var "Help"
type button
label "Help"
shell "$BIRCH/script/gde_help_viewer.csh $BIRCH/doc/bioLegato/blrevcomp.html"
close false
This aspect of BioLegato is what sets it apart from any other program of its kind. Because any program on the system can be called, there are few limits to what BioLegato can be asked to do. As well, the simplicity of the .blmenu files means that it is easy to add new programs to BioLegato, and that the cycle of testing and modification is far more rapid than would be required if it was necessary to re-compile BioLegato every time a program was added.
A complete description of syntax for .blmenu files is found in
The Command layer - The command built by BioLegato is executed by the Unix shell. If the command is run in the background (not shown in the example above) then it will run independently of BioLegato, which is especially important for long-running jobs. In fact, once the job has been started, the BioLegato window that launched it can be exited, with no effect on the job. BioLegato can call anything that can be executed, either a binary, a Unix command, a script, or anything that is executable. In a certain sense, BioLegato can do anything that a person can do at the command line.
The Computation layer - Because the command(s) executed by BioLegato can include scripts, it is possible to launch scripts that orchestrate long computational pipelines involving many programs. There is not even any requirement that processing occur on the same machine that BioLegato is running on. In another example, the BLAST menus in the default BIRCH distribution call the NCBI blastp client, which sends BLAST searches to NCBI. Except for the time required to get the results back, it is transparent to the user where the searches are run.
|
|
    |
|