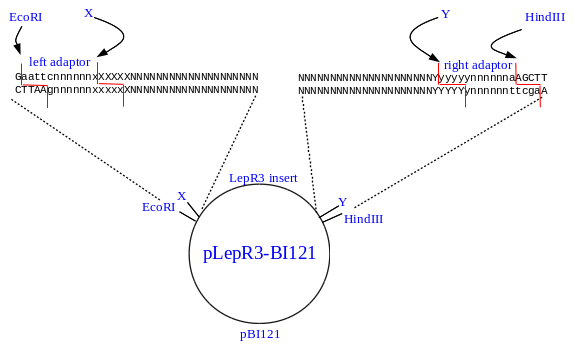
UPPERCASE - nucleotides from pBI121 or from the PCR primers
lowercase - nucleotides from the adaptors
NNNN - nucleotides from original primers
XXXXXX, YYYYYY - recognition sequences for enzymes X and Y
nnnnnn - arbitrarily chosen nucleotides for primers or adaptors
Restriction cuts generating sticky ends are shown as red lines.