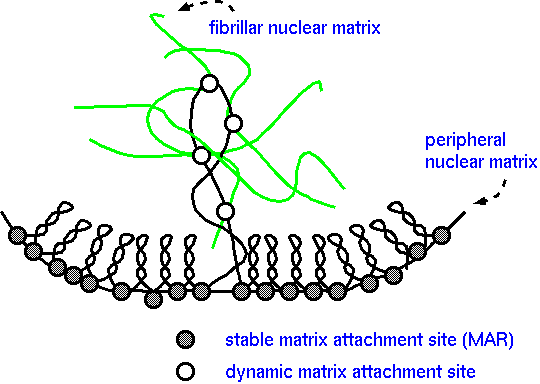
| Chromatin
organized into loop domains by stable attachment to the
nuclear matrix at approximately 50,000 base pair
intervals. Most domains are condensed into higher order
chromatin structures. The DNA of active domains is
extended by multiple sequence-specific dynamic
associations with the nuclear matrix. |
 |
There is good evidence that DNA replication and transcription of genes takes place primarily in regions in contact with the internal matrix. In this model, DNA would be threaded through the matrix attachment sites, until the appropriate gene or origin of replication was found. Then replication complexes or transcription complexes would open up the chromatin further, and carry out their functions.
Each domain can be independently regulated. To be transcriptionally active, a domain must be extended (partly uncoiled) into the fibrillar nuclear matrix. Domains that remain coiled are clustered at the periphery of the nucleus. These domains remain transcriptionally inactive. Extended domains are potentially active, but require further developmental or environmental signals to turn on transcription.
Breyne, P., Van Montagu,
M., Depicker, A. and Gheysen, G. (1992) Characterization of a
plant scaffold attachment region in a DNA fragment that
normalizes transgene expression in tobacco. Plant Cell
4:463-471.
From the previous topic, we know that chromatin
has to be in a transcriptionally "open" conformation in order to
be expressed. We know what this means in the context of
nucleosomes, but what about MARs? More specifically: What
is the effect of MARs on gene expression? Before we
look at some experimental evidence, let's set out our hypotheses:
| In an experiment to test the effect of MARs
on gene expression, tobacco cells were transformed with
several constructs. |
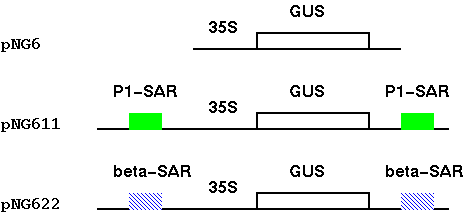 |
| Tobacco cells grown in culture as callose
(undifferentiated) tissues,
and assayed using a
colorimetric GUS assay (GUS converts the substrate
X-gluc into a blue dye). What the results
showed is that pNG611 and pNG622, which both had MARs,
showed higher expression. PNG6, which had no MARs, had a
high percentage of tissues with little or no activity. The histogram shows that tissue transformed with either the 35S-GUS gene alone, or 35S-GUS gene plus beta-gobin SAR, vary in GUS activity over a wide range. Notably, pNG6, which has no SARs, has a high percentage of calli with little or no activity. In contrast, calli transformed with GUS + P1-SAR had GUS activities over a more narrow range, with 75% of the transformants falling into a narrow window between 20 and 80 Units of enzyme/mg total protein. |
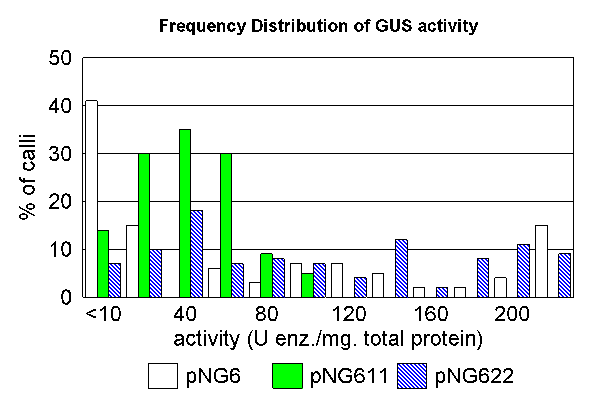 |
What this means is that the presence of MARs flanking a gene
appear to create a chromatin domain, leading to a reproducable
level of expression in independent transformants. Without the
MARs, expression of a transformed gene is less predictable.
Sometimes the gene will be inserted between two compatible MARs,
and sometimes it will be inserted in a site that is unfavorable
for expression.
|
It is likely that MARs are
necessary and sufficient to define chromatin domains.
That is, all you need to create a domain is two flanking
MARs. Other experiments have shown that a single MAR is
inadequate to confer reproducible expression in plants.
|
|
| Topoisomerases have been found to be associated with MARS. By balancing the activities of the topoisomerases, the cell can regulate the degree of supercoiling in any chromatin domain, independently of the adjacent domains. |
|
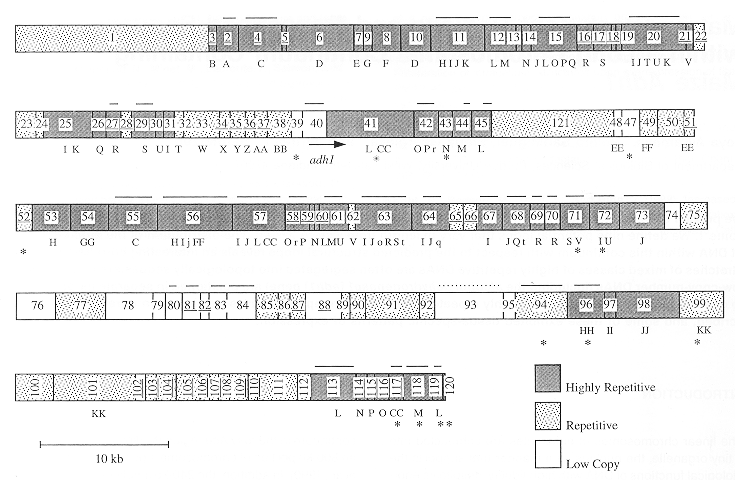
| The figure at right illustrates one
experimental strategy. If the restriction map of a region of
the chromosome is known, bands containing MARs can be
subtracted from the digest by binding to purified nuclear
matrix proteins. First, a cloned fragment from the region to be studied is digested with one or more restriction enzymes. Next, a matrix protein complex purified from nuclei, is added to the mix. Only DNA fragments containing MARs should be bound by the matrix. Upon centrifugation, the matrix proteins form a pellet at the bottom of the tube, carrying any MAR-containing fragments. The only restriction fragments remaining in the supernatant should be those without MARs. The pellet is resuspended with a detergent to break up DNA/protein complexes, and loaded onto a gel next to a lane containing the original restriction digest |
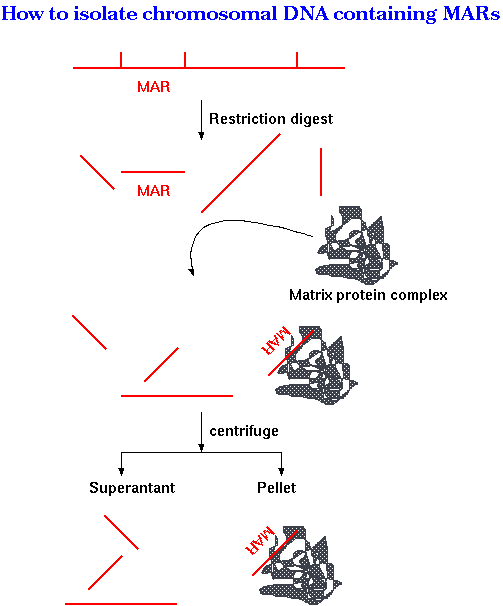 |
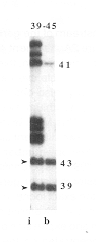
The gel at right shows the restriction
digestion of the region including fragments 39 through 45,
from the map below. Labeled insert DNA was digested with
XbaI, XhoI and Bsu36I.
Arvramova Z et al. (1995) Matrix attachment regions and transcribed sequences within a long chromosomal continuum containing maize Adh1. Plant Cell 7: 1667-1680. |
 |
What these results tell us:
Historically, cytogeneticists observed what appeared to be a
protein scaffold underlying chromosome structure. The sites of
attachment of chromatin to the scaffold were referred to as
scaffold attachment regions (SARs). We now know that SARs are the
same sites of attachment as MARs. Evidence indicates that the
scaffold (which is the basis for chromosome condensation) is
derived from the matrix in prophase, and presumably the scaffold
helps in the re-formation of the matrix at telophase.
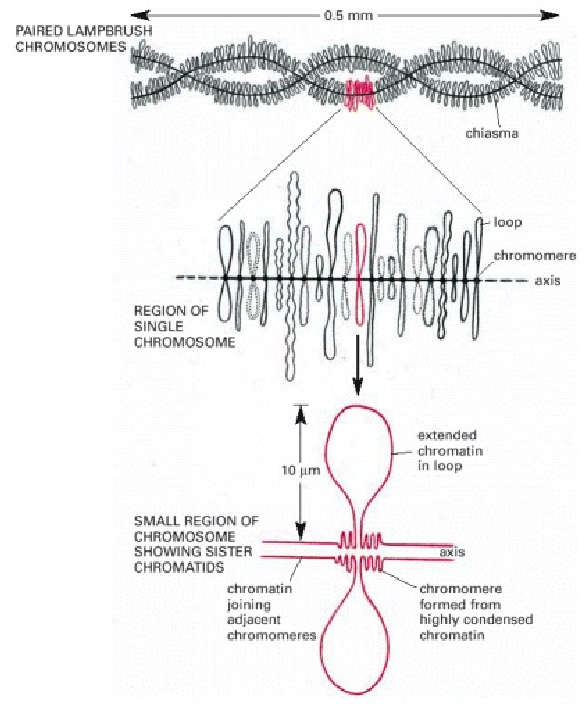
| MARs seem to be consistent within individuals
of one species. In salamander oocytes, meiosis is arrested in meiotic metaphase I and the chromatin becomes extended, allowing transcription to resume. Oocytes build up a supply of mRNA in this fashion. The extended appearance of the chromatin on these chromosomes has led to the name "lampbrush" chromosomes. The symmetry of lampbrush chromosomes provides evidence that attachment of chromatin loops to the scaffold is not random. |
 |
| It has been estimated that the set of
lampbrush chromosomes contains a total of about 10,000
different chromatin loops in many amphibians, with the
remainder of the DNA being highly condensed in the
chromosomes. Note that each loop corresponds to a particular
DNA sequence, and that four copies of each loop are present
in each cell, since the structure shown at the top consists
of two paired homologous chromosomes and each chromosome is
composed of two closely apposed sister chromatids. This
four-stranded structure is characteristic of this stage of
development of the oocyte (the diplotene stage of meiosis).
i. First, within a given oocyte, the "lampbrush" pattern of loops appears symmetrical, suggesting that the loops are the same in both chromatids. The loop pattern is also constant from one oocyte to the next and from one individual to the next. ii. Secondly, when 3H-RNA probes of specific genes are hybridized in-situ to "lampbrush" chromosomes, a given probe always hybridizes to the same loop. |
 Displayed by direct hypertext link to Alberts et al., Molecular Biology of the Cell http://www.ncbi.nlm.nih.gov/books/NBK26847/figure/A653/?report=objectonly |
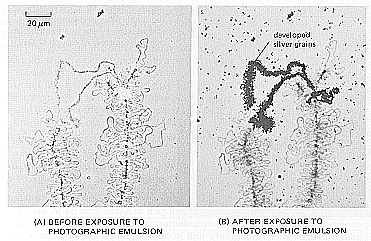
| A
single-stranded DNA radiolabled probe was prepared,
corresponding to a repeated DNA sequence containing
histone genes. The chromosomes in (A) were annealed with
this probe, washed extensively, and then subjected to
autoradiography (B). The extended loop that becomes
radioactive here is synthesizing unusually long RNA
transcripts that contain copies of several clustered
histone genes. The fact that these long RNA transcripts
hybridize with the DNA probe reveals that the repeated DNA
sequence of the probe is copied into RNA, even though in
other cells this sequence serves as a nontranscribed
spacer between the histone genes. Incidentally, this is an extraordinary case of transcription during meiosis. |
 From M.O. Diaz, G. Barsacchi-Pilone, K.A. Mahon, and J. Gall, Cell 24:649-659, 1981. |