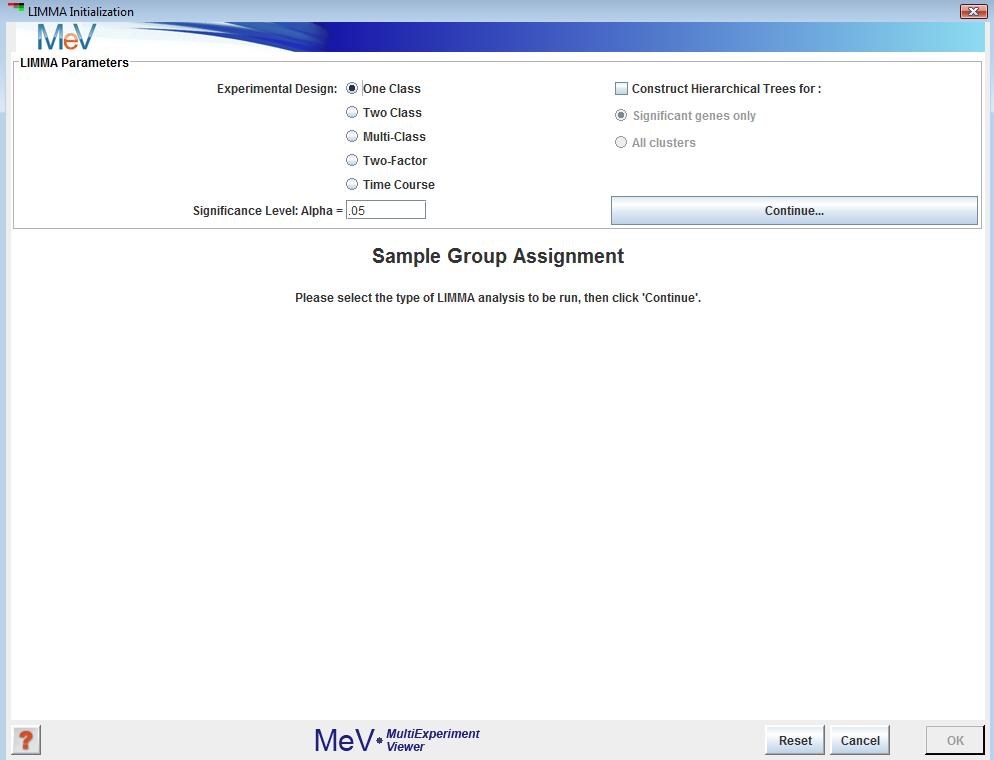
LIMMA Intialization Dialog
(Smyth, G. K. 2004)
LIMMA, a new module as of MeV 4.5, is a technique for identifying differentially expressed genes based off of the fitting of each gene to a linear model. This method can be applied to a number of experimental designs such as single class, two or more groups, as well as factorial and time-course experiments.
After opening the LIMMA initialization dialog, select the type of data you intend to run the analysis on. Your options are:
Enter the number of time points in your data.
Assign a significance level
Genes with a p-value less than alpha will be deemed significant. Click “Continue” to move to the next step.
Assign your samples
Depending on the type of data you have selected, you will need to assign your samples to groups, factors or time-points, if necessary.
Button Selection
Use this option if you have not created clusters for your samples and want to assign each sample individually.
Each group must contain at least two samples assigned to it. For factorial and time-course data, each combination of levels in factor A and factor B must contain at least 2 samples.
For example: if an experiment has 3 levels for factor “age” and 2 levels for factor “sex” . There must be at least two samples in each of the following:
Cluster Selection
Use the cluster selection panel if you have previously made clusters and wish to run your analysis based on those clusters. Use the drop-down boxes to choose which clusters’ samples are to be assigned to which time-points. For clusters you are not using, leave “unassigned”. The same condition and time-point requirements exist for this method, but the cluster selector makes for easier and more organized sample assignment.
Hierarchical Clustering
To have LIMMA construct hierarchical trees for your results, check the corresponding check box. Select whether this feature is to be applied to significant genes or significant and non-significant genes. This process may add significantly to the computation time.
The LIMMA module outputs standard viewers and tables for MeV’s statistics analyses.