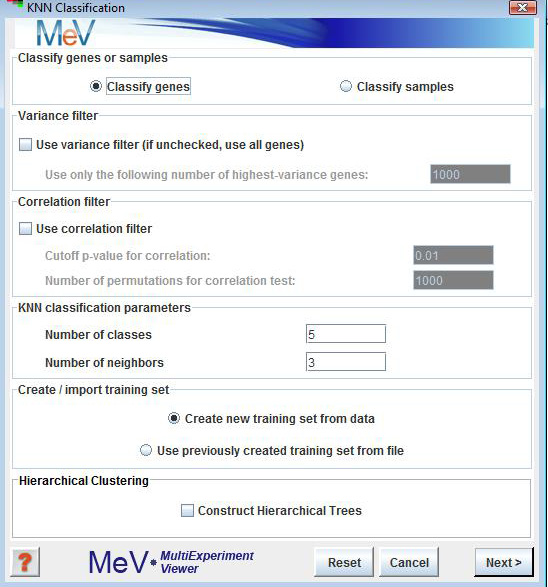
KNN Classification Initialization Dialog
(Theilhaber et al. 2002)
KNN Classification is a supervised classification scheme. A subset of the entire data set (called the training set), for which the user specifies class assignments, is used as input to classify the remaining members of the data set. The user specifies the number of expected classes, and the training set should contain examples of each class. If the option is chosen from the initial dialog, an input dialog box is displayed for parameter input. You can classify unknowns based on a training set consisting of knowns. You can validate the training set by leaving out each element of the training set in turn, and using the remaining elements of the training set to classify the one left out. You can thus assess the quality of the training classes. A good training class should have most of its original members classified into itself during this process (cross validation), and it should have few members of other training classes falsely assigned to it.
Classify genes or samples
This is self-explanatory. Although the following description refers to genes, the same steps will apply to experiments if “Classify samples” is chosen.
Variance filter
This is the first of two noise-reduction filters that can optionally be applied before classification. The variance filter keeps only those genes in the entire data set (including the classifier set) that have the highest variance across all samples. The number of genes to be retained is specified by the user.
Correlation filter
The correlation filter is used to filter out those genes of the set to be classified, that are not significantly correlated with at least one member of the training set. The significance of correlation is determined by the p-value, which is calculated by a permutation test in which each gene is permuted a user-specified number of times.
This is where the user specifies the expected number of classes (which is also the number of classes present in the training set).
The number of neighbors is the number of genes from the training set that are chosen as neighbors to a given gene. Euclidean distance is used to determine the neighborhood. Let’s say we want to classify a gene g. Gene g is assigned to the class that is most frequently represented among its k nearest neighbors from the training set (where k is specified by the user). In case of a tie, gene g remains unassigned.
Create/import training set
If the user chooses to import a previously created training set (for instructions on saving a training set, see below), on hitting the “Next” button a file chooser is displayed from which the training file can be chosen. If an appropriate file is chosen, the KNN classification editor shown below in Fig xx is displayed with the class assignments from the file. If the option to create a new training set from data is chosen, on hitting the “Next” button the classification editor is directly displayed with all genes set to neutral.
Hierarchical Clustering
This checkbox selects whether to perform hierarchical clustering on the elements in each cluster created.
Genes can be assigned to any of the specified classes by checking the box in the appropriate column for that gene. Genes designated as “Neutral” will be classified in subsequent steps, whereas those assigned to classes in this step will be treated as the training set. The “Edit” and “Tools” menus at the top of the classification editor allow searching, sorting and selection of data. The classification scheme can be saved in a text file if needed, to be loaded in a future KNNC run (as explained above).
On hitting the “Next” button in the classification editor, the calculations proceed, and the output has the usual viewers as most other MeV runs. The main difference is that each type of viewer has several sub-viewers for Used Classifiers, Unused Classifiers (those that are possibly weeded out by variance filtering), Classified, Used Classifiers + Classified, Unclassified and All. This is done so that users can visually compare training vectors to trained vectors.
The option in KNNC provides dialogs very much like the above, and is used to perform leave-one-out cross validation on the training set. The output viewers are also similar to the ones obtained in classification, with an additional viewer that provides cross-validation statistics.