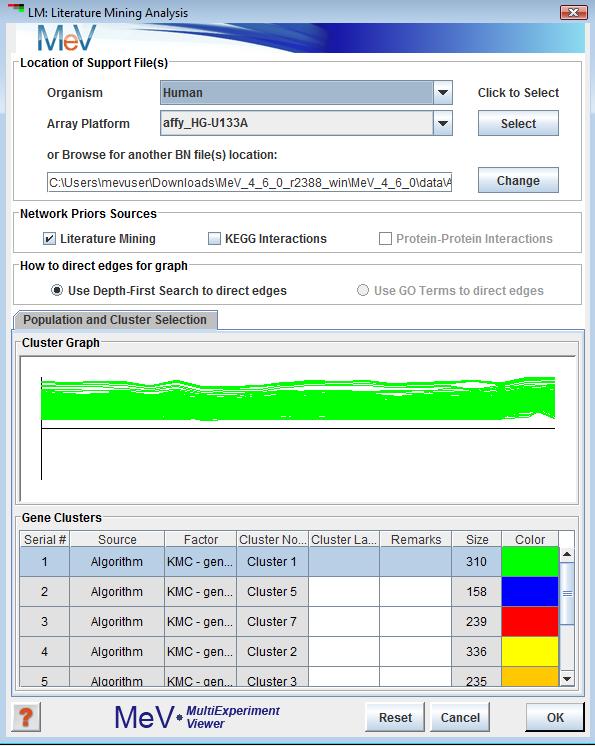
LM Intialization Dialog
(Djebbari and Quackenbush, 2008)
Selection this analysis will display the following window. This window collects all the parameters and inputs required to run the analysis. All the options in this dialog are explained below.
This option allows users to select the location where all support files needed to run BN. A description of files required can be found (see appendix for BN file descriptions).
The checkboxes provide the users to select the source of Bayesian prior probabilities in constructing a seeded network. Currently Literature Mining and KEGG priors are available. The Protein - Protein Interaction as a source of priors is still under development.
As of now, the KEGG support files are automatically downloaded from TN4 website by the application. The user is prompted for Species information if annotation is not available. All other prior sources must be made available.
The data mining algorithm requires that the data be discretized into bins before it can be evaluated for network structure learning.
It is strongly recommended that user selects the default value of 3, which means the data can exist in 3 states:
Under expressed
Over expressed
Unchanged
The algorithm functions and reports meaningfully if the 3 state rule is followed.
The algorithm uses DFS or Depth First Search to connect nodes in the initial seeded network. For large networks with lots of nodes this can take a while to complete. The GO Term option of directing edges is not yet fully developed.
The input cluster on which the analysis runs. If the user has saved more than one cluster, one of those clusters must be selected from the clusters panel. By default the first is selected.
The LM viewer displays the network file names and locations that were created during the analysis. On Right Click it displays a popup menu to launch Cytoscape via Webstart with all files created.
All result files are stored in the Where {..} represents the directory where the BN/LIT analysis supporting files are located.
Once the analysis completes it automatically launches Cytoscape Cytoscape via Webstart with the result networks displayed in it.
We have pre-created support files needed to run BN or LM analysis for some popular microarray platforms like Affymetrix, Agilent etc. Currently we are providing support files 3 species Human, Mouse & Rat. MeV comes preloaded with the files for 2 array types in the ~/data/BN_files folder.