Moving bases by hand
There are two tools available for moving bases by hand. One is the Move Blocks tool (![]() ), the other is the Push Sequence tool (
), the other is the Push Sequence tool (![]() ).
).
Both of these tools have a "Live update analyses, etc." option in their popup menus. If this is chosen, then all calculations (e.g., charts, character tracings, etc.) that depend upon these data will be redone as you use these tools. (Recalculations will be done once you let go of the mouse button, in any event.) If some of these calculations take a while, this can dramatically slow down use of these tools.
There are some visualization features in Mesquite that can aid in manual alignment.
 Move Blocks Tool
Move Blocks Tool
This tool will move a contiguous block of a single sequence. If you move the tool between two bases it will let you split a block in two, inserting gaps.
If you wish, you can move more than a single, contiguous block by turning on "Move Selected Block" within the tool's popup menu, then using the tool to move selected bases and gaps.
 Multiple Block Splitter
Multiple Block Splitter
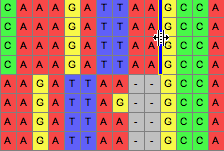
This tool allows one to draw a line between two sites, through one or more sequences, as shown in the following image:
Then, if the tool use used upon the line, those sequences can be dragged, opening up gaps along the line:
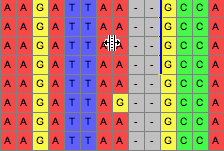
If you hold the Option key down, then all sequences other than those delimited will be moved. There are additional options in the popup menu of the tool's button that allow you to request that entire sequences are moved, or all sequences on one side. You can also request that the matrix be expanded on either end as necessary as you move sequences around; by default this option is off.
 Selected Block Mover
Selected Block Mover
This tool move move a selected block of cells. There are additional options in the popup menu of the tool's button. For example, you can also request that the matrix be expanded on either end as necessary as you move sequences around; by default this option is off.
 Push Sequence Tool
Push Sequence Tool
This tool pushes to the right the entire sequence to the right of the tool, or, if you have the Option key held down, it pushes to the left the entire sequence to the left of the tool.
Shifting sequences
Select a portion of a single sequence. If you then choose Matrix>Alter/Transform>Shift Other to Match..., then you will be presented with a dialog box that lets you choose a range of taxa, as well as a percentage. By default the range of taxa is the remaining taxa in the matrix. The default percentage is 75%.
If you press OK, Mesquite will look at each of the taxa within the range, and see if it can find a portion of each that is at least as similar as the percentage indicated to the selected portion (no gaps allowed). For each taxon that it can find such a match, it will shift those sequence such that the matched regions are lined up with the selected region.
Note that this is not an alignment algorithm; it simply shifts the sequences, maintaining whatever gaps are there or not in the sequences to be shifted.
You can also choose Matrix>Alter/Transform>Shift To Minimize Stop
Codons to shifts each
sequence 0, 1, or 2 bases so as to minimize the number of stop codons. The amount
each sequence will be shifted will vary from sequence to sequence. This feature
requires that codon
positions be designated.
Removal of Gaps
Remove All-Gap Characters
Choosing this from Matrix>Alter/Transform>Remove All-Gap Characters will remove all sites in the matrix that consist entirely of gaps.
Collapse Gaps
With a block of the matrix selected, choosing Matrix>Alter/Transform>Collapse Gaps will remove all gaps in the selected block, with the bases or amino acids pushed as far to the left as possible.
Selecting Hint
To select from one cell to the end of the sequence, select the cell and then hit Shift-Alt-Arrow key (or, on OS X, Shift-Option-Arrow key), where the arrow key is either the right or left arrow key.
Preparing the Matrix
Several options under Matrix>Alter/Transform may be useful to modify the matrix in preparation for alignment. Some of these options are under "Other Choices...".
- Collapse Gaps to Left
- Collapse Gaps to Right
- Trim Terminal Gaps Character
- Gaps to Missing
- Missing to Gaps
- Terminal Gaps to ?
- Terminal ? to Gaps
- Reverse Complement
- Reverse
- Nucleotide Complement
