BIRCH
is
- is a comprehensive
desktop bioinformatics system integrating
many of
the commonly-used bioinformatics programs
- is a
framework
of tools, files, and documentation for organizing
and managing a bioinformatics
core
facility
- can
be customized by seamlessly merging 3rd party
applications into BIRCH
|
 |
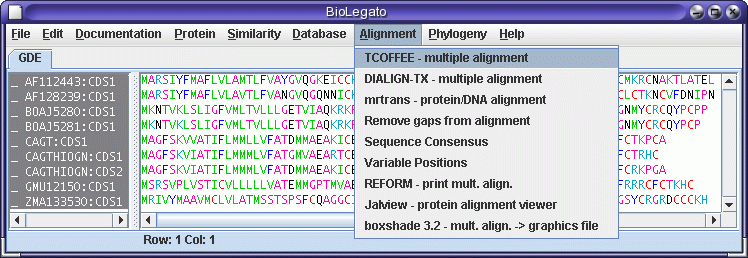
| BioLegato
is
best thought of as a program that runs other programs.
bioLegato is the
primary graphic interface for launching programs in
BIRCH. For each
program, bioLegato provides a menu that lets you set
program
parameters,
launch the program, and view the output. bioLegato takes
care of all of
the
background details, such as translating files from one
format into
another. In many cases, output also goes to a new
bioLegato window,
making it
possible to do ad
hoc
pipelining. This is one of the most powerful aspects of
bioLegato,
because it
means that you can usually run additional programs using
the output of
one program as the input of the next. In contrast, most
browser-based
applications display output in a human-readable form
that allows no
further analysis by other programs. |
 |
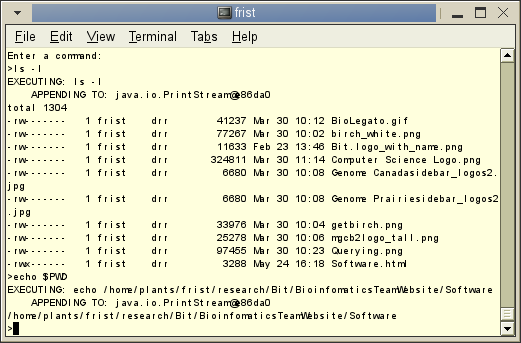
Turtleshell - Turtleshell is
a subset of the bash shell, written in Java. The big
question is, why?
- run
existing
shell scripts on any platform
- cross-platform
wrappers and scripts
Turtleshell is currently a partly-working prototype. Our
immediate goal
is to implement enough bash-like functionality to make
it possible for
BIRCH to run on MS-Windows. However, Turtleshell quite
generic, and
could potentially be used for almost any software
development project,
we
|
|
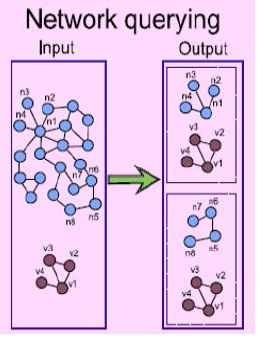
| Ph.D.
student Abiel
Roche (Computer Science) is investigating the
application of a class of
Finite State Automata, called Transducers, to the
comparison of
biological networks between different species, or
between members of
the same species in different experimental conditions.
The goal is to
develop metrics for quantifying the differences between
congruent
pathways, and for identifying subnetworks that are
congruent between
larger networks. |
 |
