Minimum Sequences
(BMC Bioinformtics 2001, 2:1)
- "For the most-typical
codon, the expected R:S is 1:3, >= 3 mutations required"
- "For cta or ctg, only 2 mutations required"
- For
negative selection detection, much more mutatations required (> 10)
False-positive Testing
(BMC Bioinformtics 2001, 2:1)
- Conduct simulation by generating 10 sets of 300 sequences of length 999 (EVOLVER (PAML) )
- The average branch length of each set varies in 0.1 intervals from 0.1 to 1.0
- False positive persentage is calculated and plot against average branch length
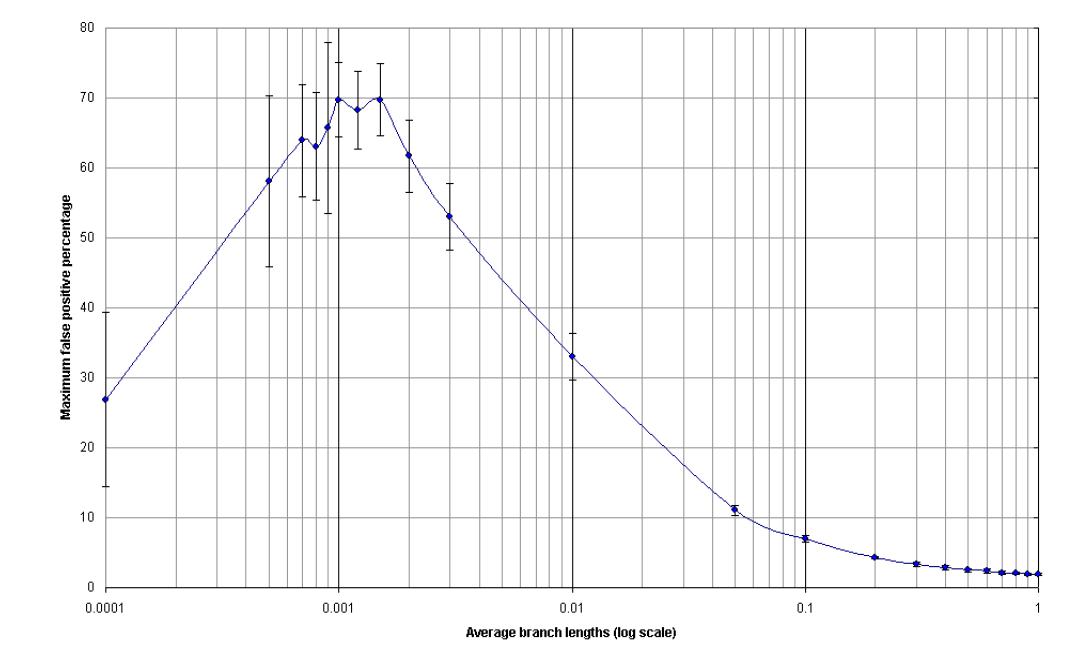
derived from Dr. Jeffrey J Stewart, 2001
False positive percentage (F) = false psoitives / (false positive + neutral drift variants)
- F <~2%, if branch lengths are in range of 0.1 --- 1.0
- F >~70%, if branch length
is at 0.001