
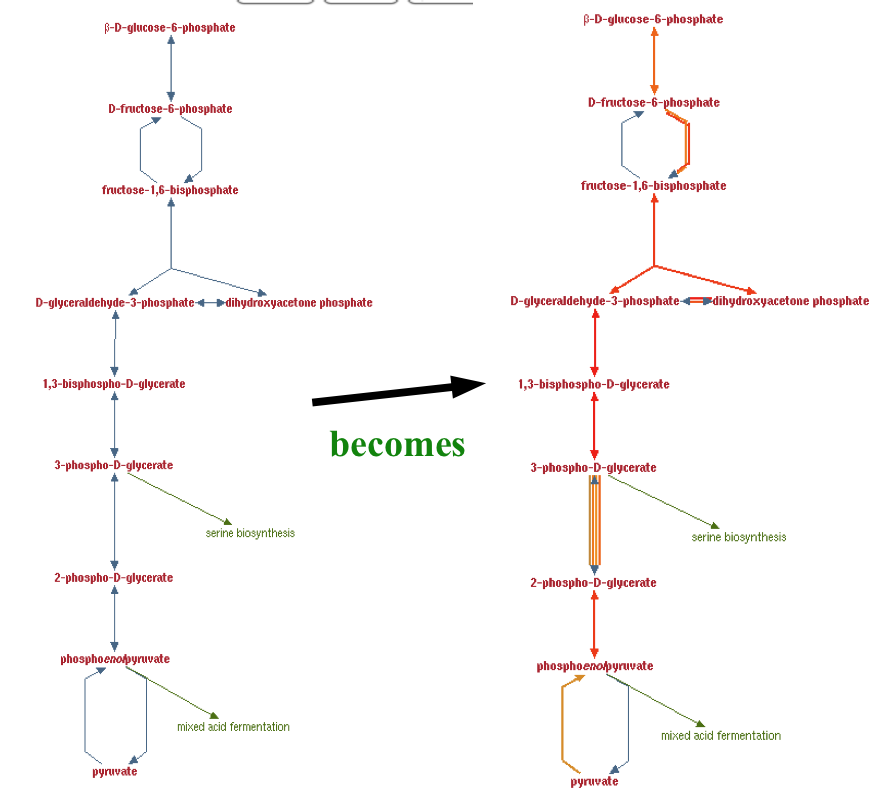

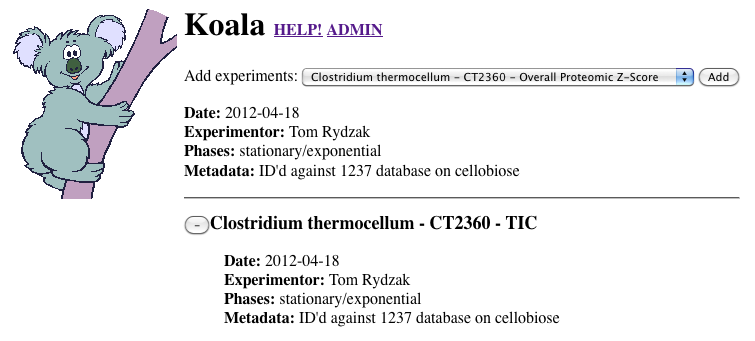
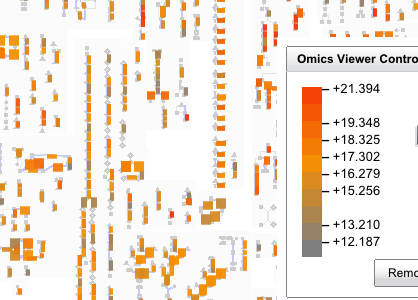

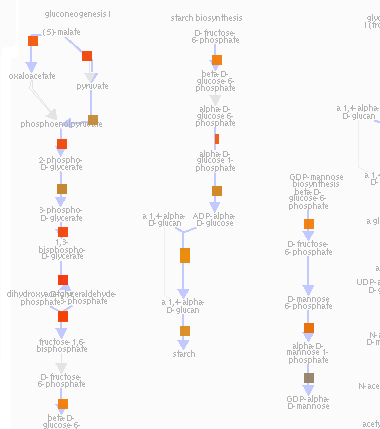
Koala is a Biocyc Pathway Tools compatible plugin, which facilitates overlaying, and storing transcriptomic and proteomic data and metadata.
Summary: Research groups often wish to visualize their proteomic, transcriptomic, and microarray data overlaid onto the corresponding metabolic pathways. The SRI-developed BioCyc Pathway Tools software suite provides many ways to do this, including Omics Viewer and Cellular Overview. Koala is a web-based BioCyc Pathway Tools-compatible plug-in designed to complement this by enabling researchers and workgroups to overlay their experiment data on any Pathway Tools visualization while sharing, storing, and managing their data. Koala also provides a system to collect and manage experimental metadata in a publication ready format.
Availability and Implementation: Koala is freely available under the GNU General Public License (GPL) v3 on the web at: http://home.cc.umanitoba.ca/~psgendb/local/koala/. Koala is implemented in HTML, PHP, Javascript, MySQL, and Apache with all major browsers supported.
Brian Fristensky
Bioinformatics Lab
Department of Plant Science
University of Manitoba
Winnipeg, Manitoba, Canada
R3T 2N2
E-Mail: frist@cc.umanitoba.ca
Webpage: http://home.cc.umanitoba.ca/~frist
Graham Alvare
Bioinformatics Lab
Department of Plant Science
University of Manitoba
Winnipeg, Manitoba, Canada
R3T 2N2
E-Mail: alvare@cc.umanitoba.ca
Webpage: http://home.cc.umanitoba.ca/~alvare