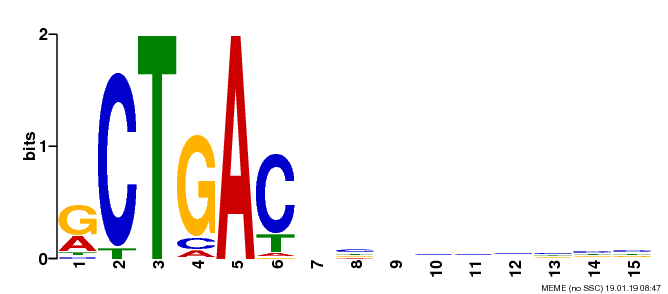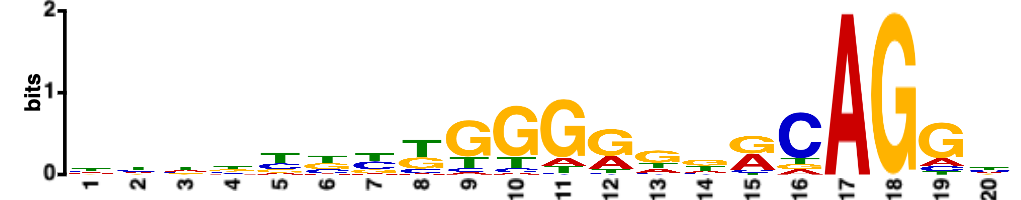|
Laboratory
of RNA Biology
|
|
Control of alternative pre-mRNA splicing in biology and diseases
Alternative splicing is a common way of gene regulation that allows the generation of multiple mRNA and often protein isoforms from a single gene. Almost all human gene transcripts are alternatively spliced and some are known to generate extremely diverse protein isoforms. This greatly contributes to the proteomic complexity, particularly in neurons and endocrine cells. In experimental animals, genetic deficiencies in alternative splicing factors result in developmental defects or embryonic-lethal phenotypes. In humans, aberrant splicing accounts for the defect of at least 30% of genetic mutations that cause diseases. Thus, it is important to understand alternative splicing and its regulation in biology and diseases in the omics era.
For
example, in electrically excitable cells such as
neurons, endocrine and muscle cells, ion channels
allow ions in/out of the cell membranes to generate
electrical firing patterns that are important for cell
functions including memory, hormone secretion and
muscle contraction. These processes are believed to be
critical for higher order phenotype such as learning,
behavior, metabolism and heart beating. How these
processes are finely tuned during development and in
adult life is still a mystery to researchers.
Alternative splicing provides a unique way to
diversify proteins and may play a critical role here.
A number of studies have indicated that alternative
splicing is involved in adaptive or addictive changes
in neurons by neuronal activity or alcohol
stimulation.
Interestingly,
alternative splicing of some ion channel genes is
regulated by membrane depolarization, the first part
of an action potential, implying a gene expression
change related to the electrophysiological
memory observed in neurons,
or hormone
production in response to experience in life (e.g. exercise or stress).
However, the molecular basis of the splicing
regulation, particularly after recurrent stimulation,
remains largely unclear.
We
have used the STREX (stress
axis-regulated
exon) variant of the Slo1 BK potassium
channel
gene as a model to study how cell signals regulate the
choice of alternative splice sites in pre-mRNA
transcripts. Inclusion of the STREX exon enhances the
calcium sensitivity of BK channels and likely
modulates cellular electrical properties related to
hearing frequency tuning or adaptive changes in
learning and memory or tolerance to alcohol. Its
regulation by stress hormones and the
calcium/calmodulin-dependent protein kinase IV (CaMK
IV) makes it an interesting target for dissecting the
components regulating alternative splicing as well as
understanding the impact of splicing regulation on
neuronal electrical properties. A first step toward
this goal was made by coupling CaMK IV with a pre-mRNA
element (CaRRE1,
Fig. 1) sufficient to confer CaMK IV response to an
otherwise non-responsive exon. We have recently
identified the splicing factors hnRNP L and L-Like (LL) as
essential components of the CaMKIV-regulated splicing
of STREX by inhibiting U2AF65 binding to the upstream
3' splice site. Particularly for hnRNP L, its Ser513 is
phosphorylated
and essential for the regulation.(Fig. 2) Other
factors including PTB and
hnRNP K are involved in the
regulation as well.
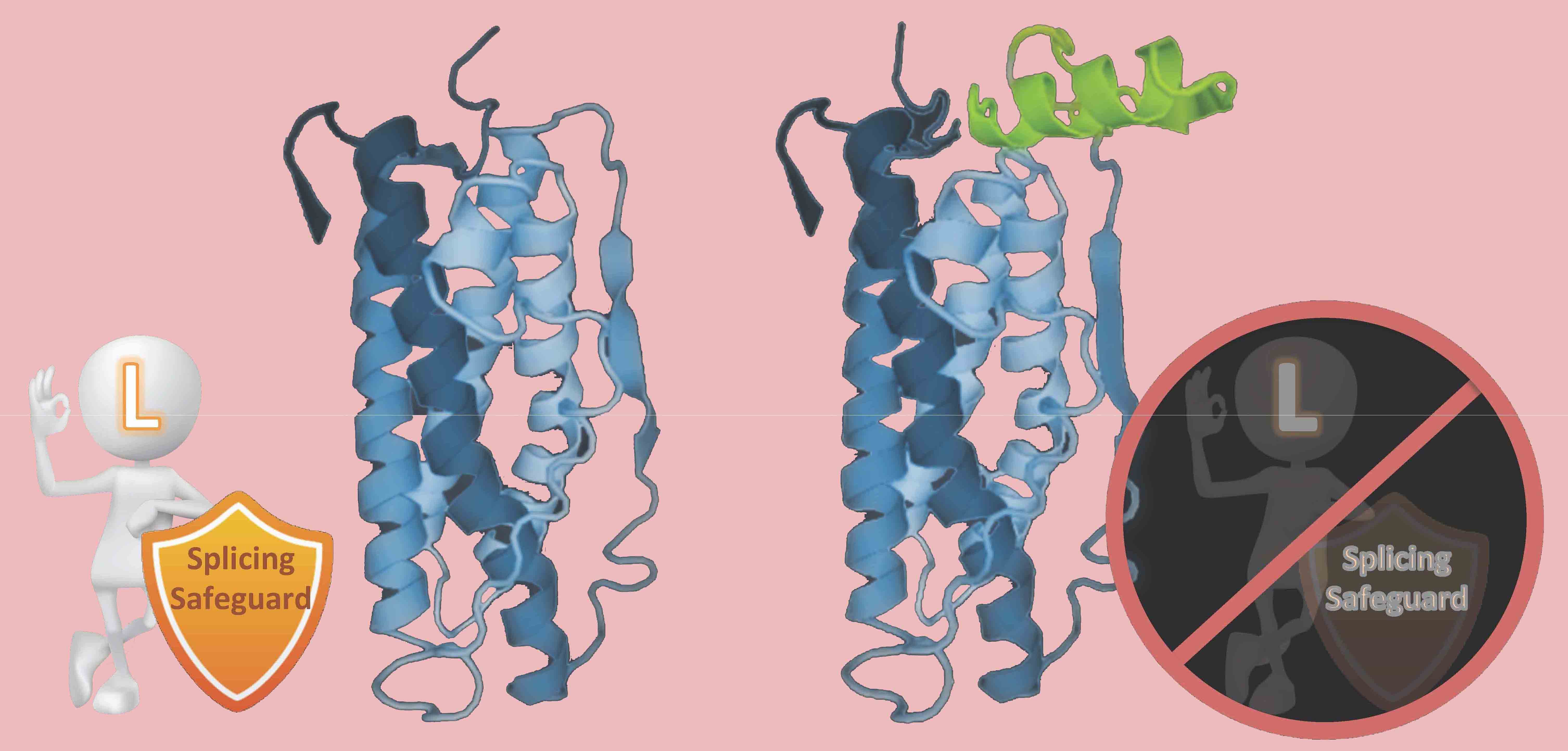
In particular, hnRNP L and LL are required for the differential regulation and protection of the hormone gene expression programs for producing prolactin and growth hormones (Fig. 3). Further detailed characterization of this molecular process and its role in homeostatic or adaptive splicing (Fig 4), hormone production and stress response is ongoing .
In
recent years, reports by several labs have also
demonstrated that alternative splicing of the BK
channel and AMPA
receptor genes after chronic inactivities or of
the neurexin
gene upon depolarization/activities of neurons
plays a critical role in the homeostasis
of cellular electrical properties or synaptic
formation. Moreover, the regulation is mediated
by the Ca++/calmodulin-dependent protein kinase IV
(CaMKIV) and its downstream splicing factors Sam68
or Nova-2,
depending on the target exons. Together, these
observations support a critical role of
depolarization-regulated splicing in hormone
production, neuronal homeostasis or development.
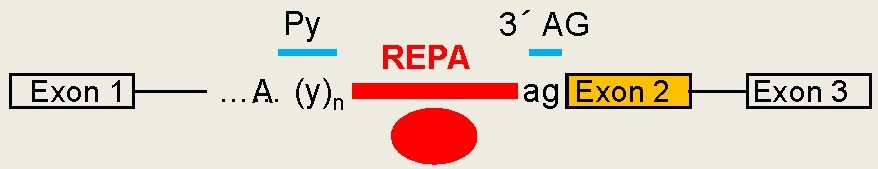
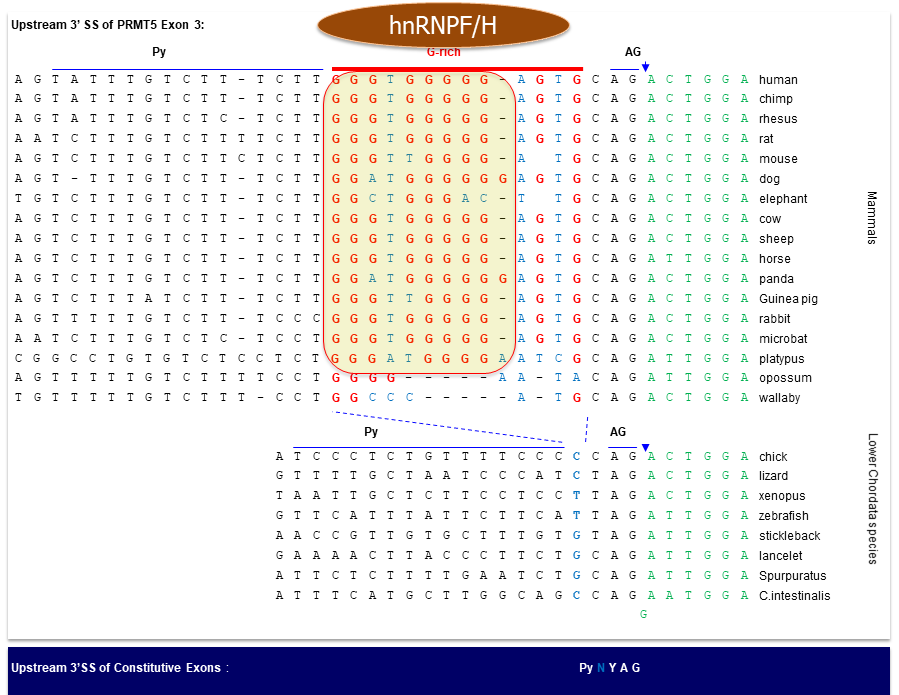
CaRRE1-like RNA
elements, even G tracts,
have been found to act similarly as CaRRE1 in hundreds
of human genes. Together we call these groups of RNA
sequences REPA (regulatory elements between
the polypyrimidine tract and 3' AG (Figs. 5 & 6). Most
of the REPA G
tracts (REPAG) appear to have
emerged in the ancestors of mammals, likely
contributing to the higher abundance of alternative
splicing and proteomic diversity. We have demonstrated
with the REPAG of PRMT5 exon 3 that it
contributes to the evolutionary emergence of a novel
splice variant with an opposite effect on cell cycle.
The REPAGs
are widespread and highly enriched in metazoa
and plants, with the highest abundance in mammals.
They are also enriched in the aberrant
3' splice sites of cancer patients mutated of
the 3' splicing factors SF3B1 or U2AF35. In the CBS gene, mutation of which causes
the human genetic disease
homocystinuria, a REPAG prevents its aberrant
splicing. In GWAS, SNPs within potential
REPA-3'SS are enriched as well, suggesting its
widespread role in curbing cryptic splicing in the
non-coding regions of the human genome.
Other
exons studied include those involved in neuronal function, human genetic disease or cell growth/apoptosis,
or splicing regulations involving other signaling
pathways including protein acetylation and methylation.
Analyses
of these signal-responsive RNA elements indicate that
they are mostly mammalian-specific, likely contributing to the more
delicate and dynamic control of alternative
splicing and higher proteomic complexity.
We
hope these studies will provide molecular details of splicing
changes in cell physiology, as well as
knowledge for cancer diagnosis/therapy or the correction of aberrant splicing
that causes human genetic diseases.
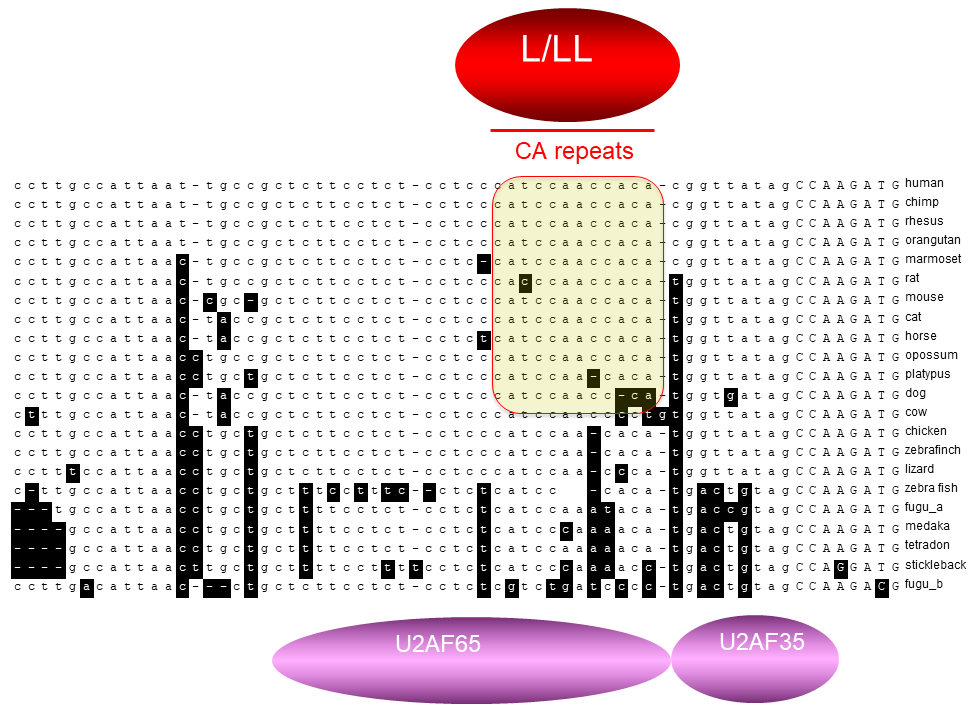
(Right-click image to view details)
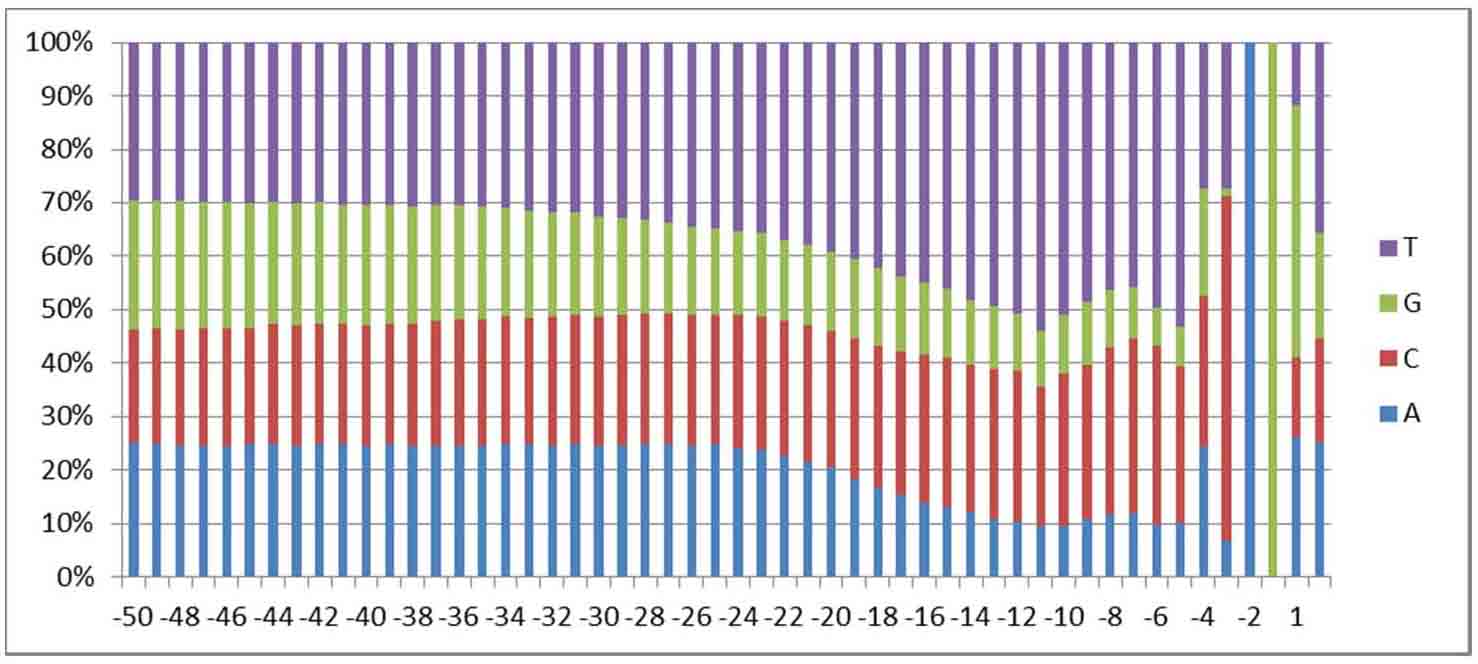
Fig. 1. The CaRRE
element among vertebrates (Liu
GD, et al., J. Biol. Chem. 2012,
287:22709–22716)
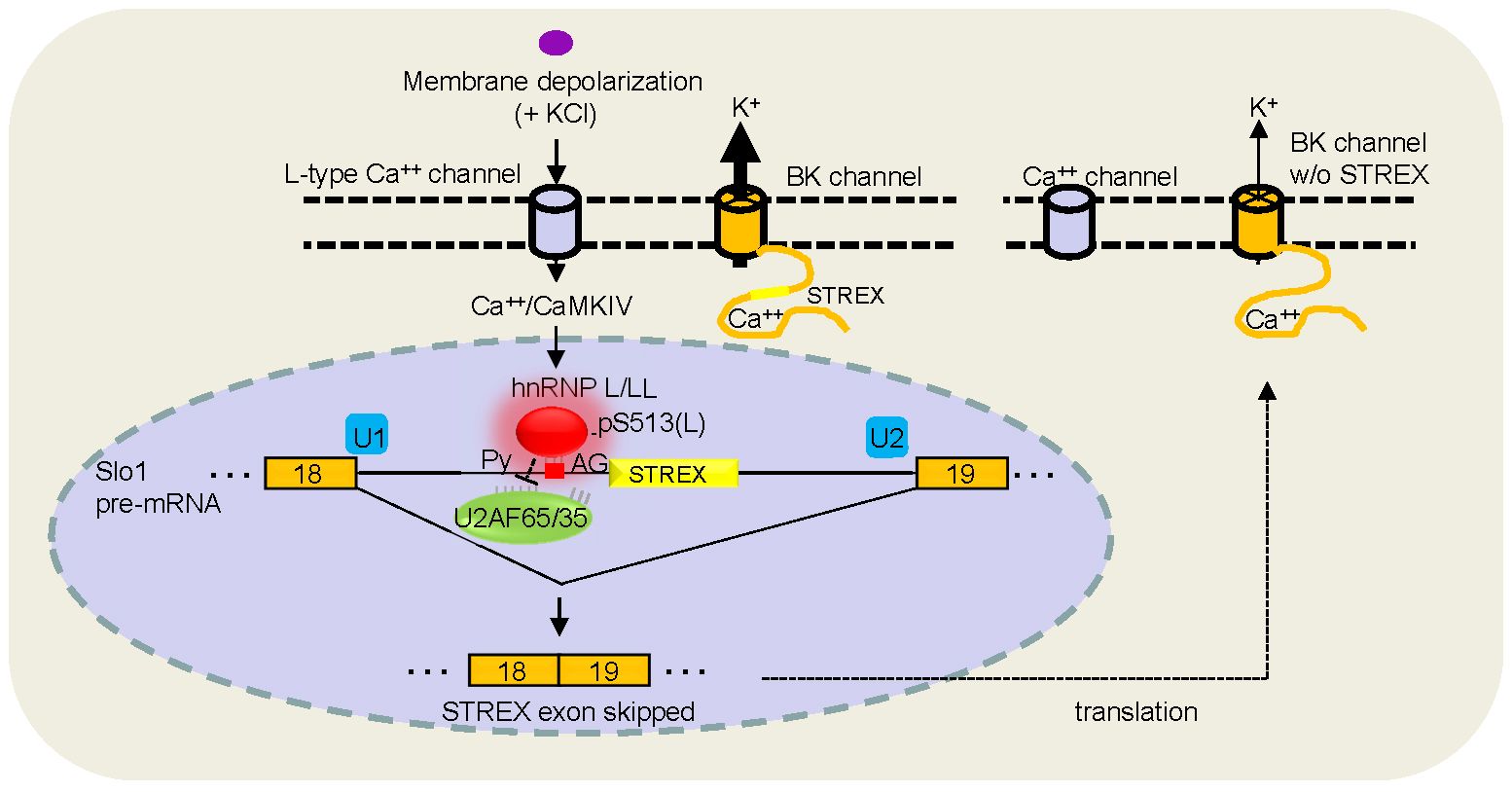 Fig. 2. Molecular basis of the
depolarization-regulated alternative splicing of the
STREX exon of the Slo1 gene.
Fig. 2. Molecular basis of the
depolarization-regulated alternative splicing of the
STREX exon of the Slo1 gene.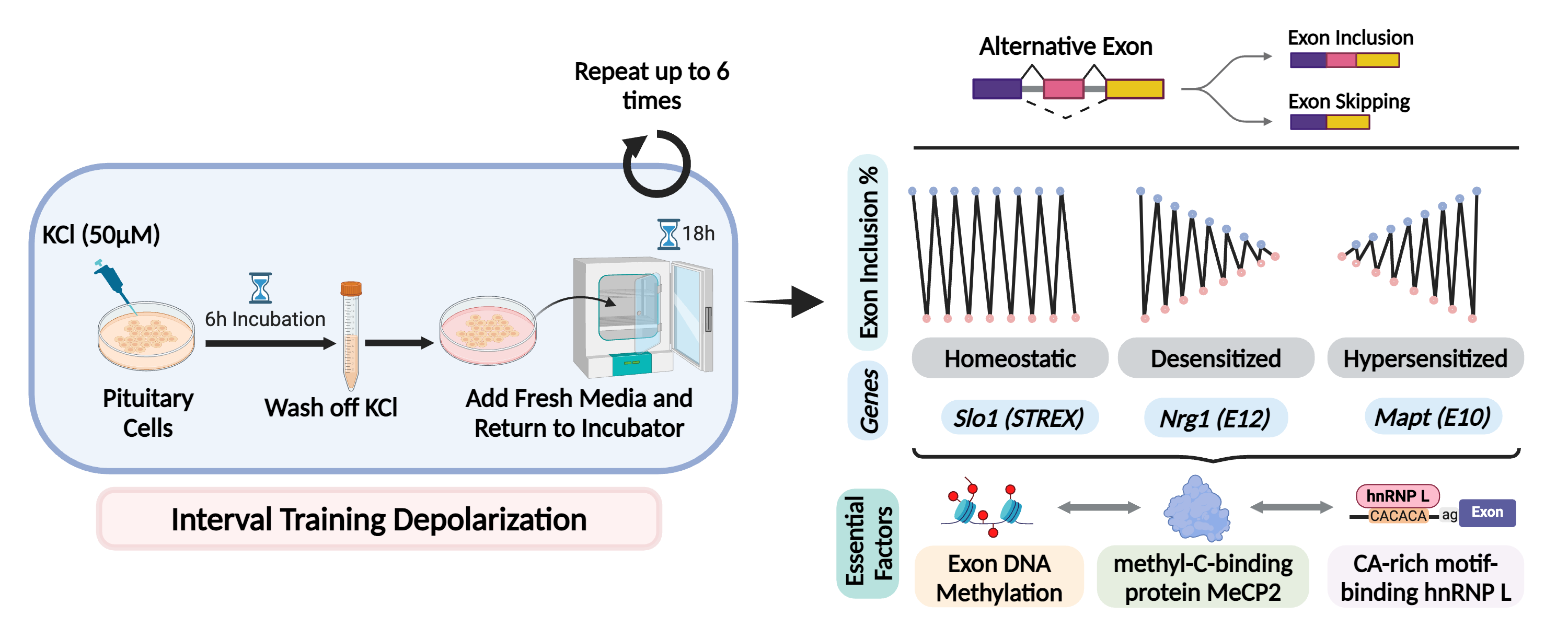
(Right-click image to view details)
Fig. 5. The
REPAG element specifically among mammals (Sohail M., and Xie J. Molecular &
Cellular Biology, 2015, 35(12):
2203-2214)
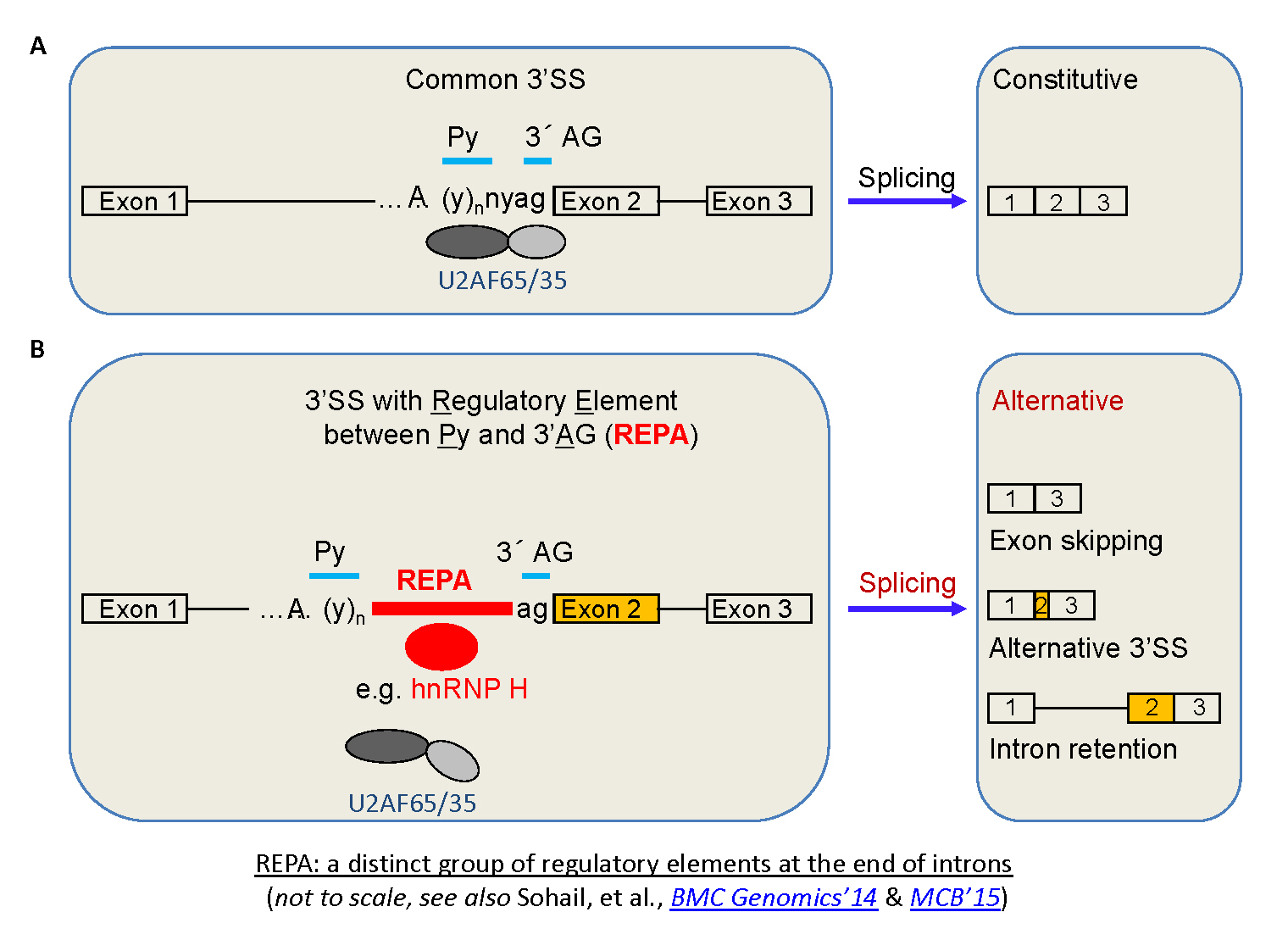
Fig. 6. A new group of introns: REPA element 'inserted' between the Py and 3'AG and its effects on alternative splicing.
IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII
Misce. links:
YOU are special - Mr. Rogers

- WES, EXON, Exome and Protein-Coding Regions, Non-coding regions, UTR
- Eradication of Polio in China
-
 RNA Meeting in Winnipeg, June
23-26, 2019 Some RNA/DNA
literature/history:
RNA Meeting in Winnipeg, June
23-26, 2019 Some RNA/DNA
literature/history: - Why are the snRNAs in splicing called U1, U2, U4, U5, U6 snRNAs? Here is the original paper that describes the use of Us in the names for some of the small nuclear RNAs that are Uridylic acid-rich, by James L. Hodnett and Harris Busch of the Baylor University College of Medicine in 1968: Hodnett JL, Busch H. Isolation and characterization of uridylic acid-rich 7 S ribonucleic acid of rat liver nuclei. J Biol Chem. 1968 Dec 25;243(24):6334-42. |PDF|
- 125 Questions in Science, which ones are related to Alternative Splicing?
- How to present a clean, highlight revision text without the deleted words in the text or balloons after tracking changes in WORD?
- Albert Einstein the mediocre: Why the h-index is a bogus measure of academic impact (Reader discretion is advised).
- Opinions
about the term
non-coding RNA