| region | (f-c)/(f+c) | ||
| A | f(2:1835)=1095 | c(2:1835)=739 | 0.194 |
| B | f(1836:3698)=1097 | c(1836:3698)=1144 | -0.228 |
Acinobacter_baumanii_MDR-TJ.fea
Acinobacter_baumanii_MDR-TJ.fea.tsv
Acinobacter_baumanii_MDR-TJ.fea.ods
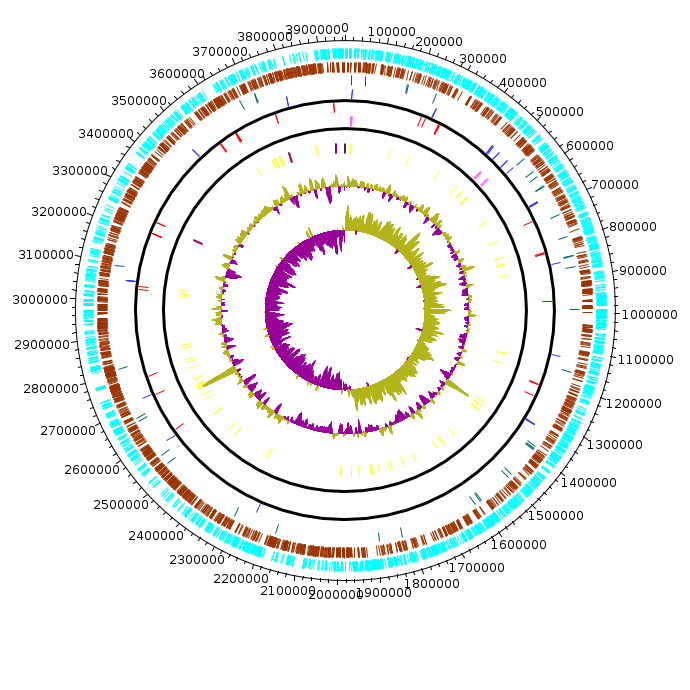
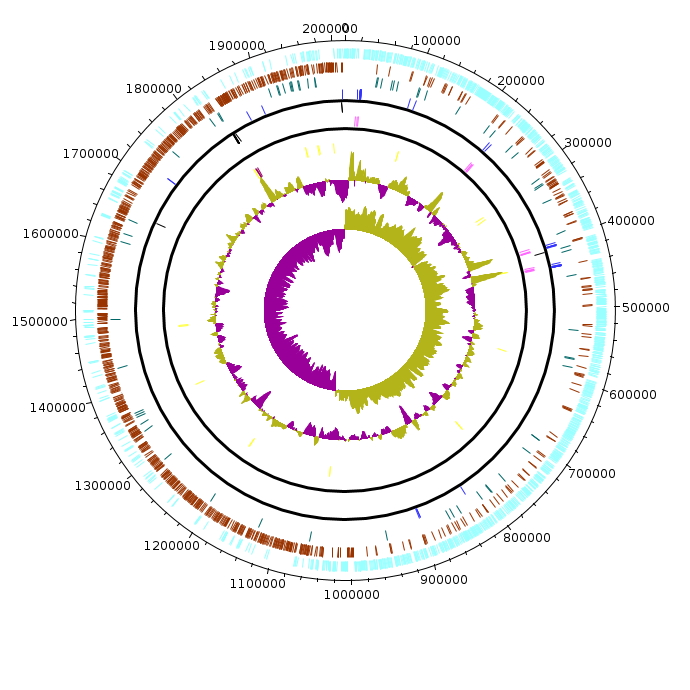
Actinobacter_baumanii_MDR-TJ
Acinobacter_baumanii_MDR-TJ.fea Acinobacter_baumanii_MDR-TJ.fea.tsv Acinobacter_baumanii_MDR-TJ.fea.ods |
 |
Candidatus_Hamiltonella_defensa_5AT
Candidatus_Hamiltonella_defensa_5AT.fea Candidatus_Hamiltonella_defensa_5AT.fea.tsv Candidatus_Hamiltonella_defensa_5AT.fea.ods |
 |
CstercDSM8
CstercDSM8.fea CstercDSM8.fea.tsv CstercDSM8.fea.ods |
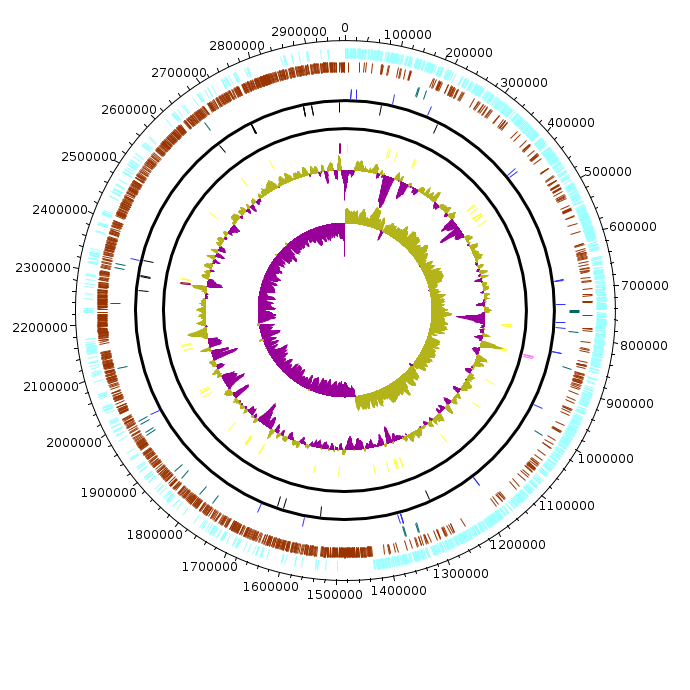 |
Escherichia_coli_synthetic
Escherichia_coli_synthetic.fea Escherichia_coli_synthetic.fea.tsv Escherichia_coli_synthetic.fea.ods |
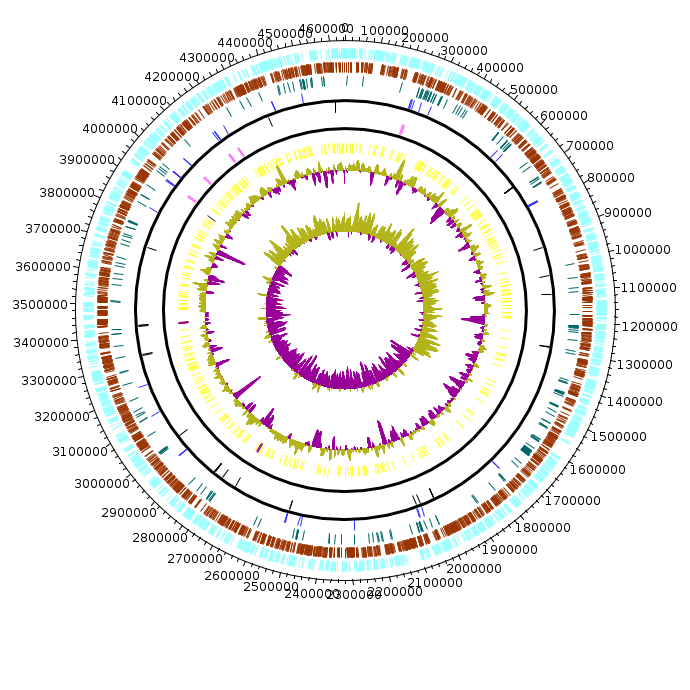 |
Halobacterium_salinarum_R1
Halobacterium_salinarum_R1.fea Halobacterium_salinarum_R1.fea.tsv Halobacterium_salinarum_R1.fea.ods |
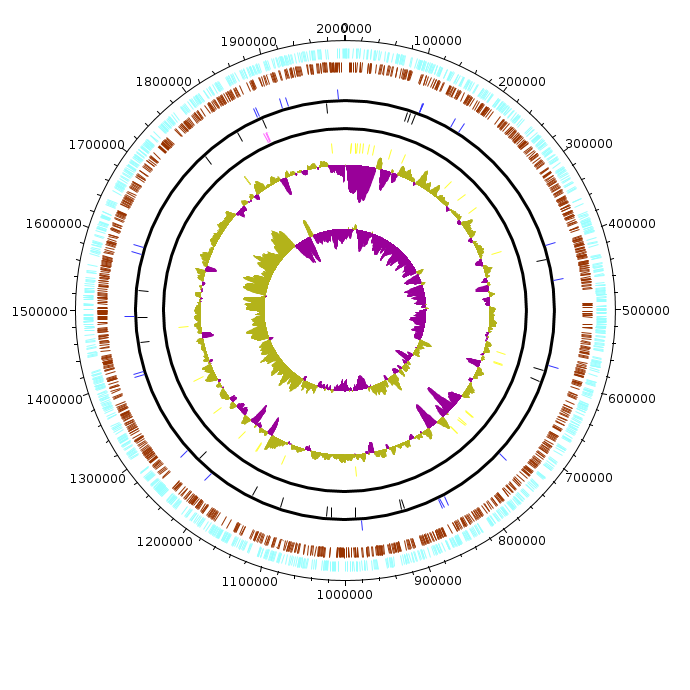 |
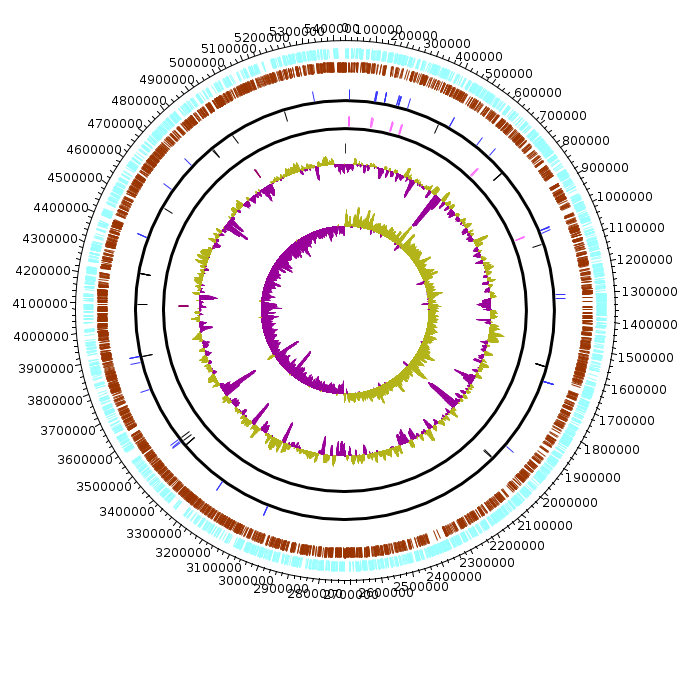
Klebsiella_pneumoniae_BAA-2146
Klebsiella_pneumoniae_BAA-2146.fea Klebsiella_pneumoniae_BAA-2146.fea.tsv Klebsiella_pneumoniae_BAA-2146.fea.ods |
 |
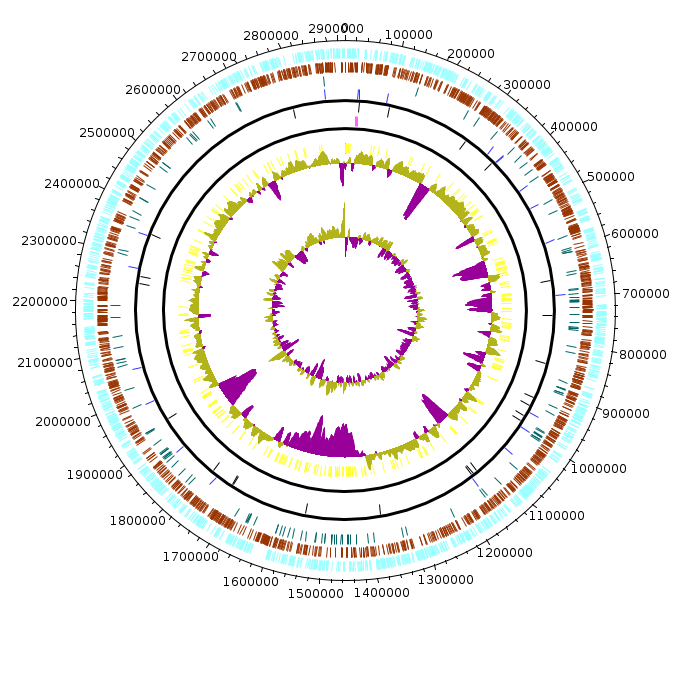
Natronomonas_moolapensis_8.8.11
Natronomonas_moolapensis_8.8.11.fea Natronomonas_moolapensis_8.8.11.fea.tsv Natronomonas_moolapensis_8.8.11.fea.ods |
 |
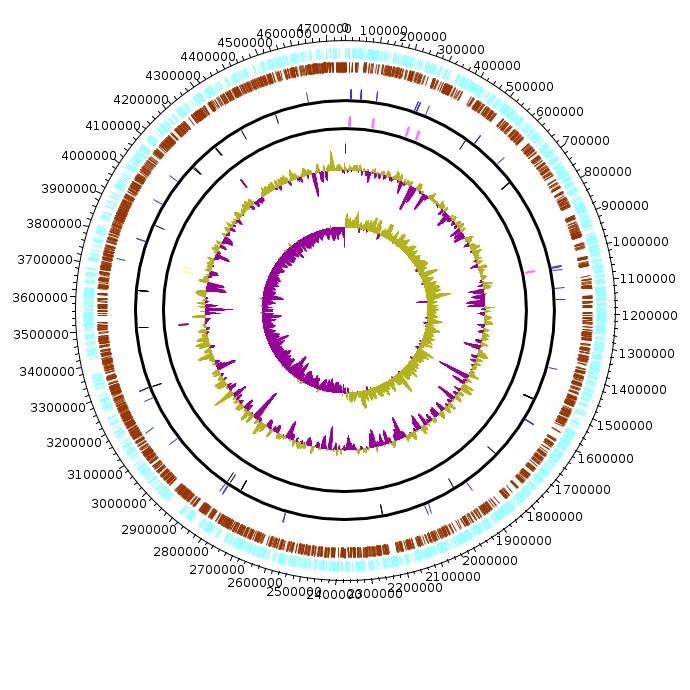
Salmonella_enterica_enterica
Salmonella_enterica_enterica.fea Salmonella_enterica_enterica.fea.tsv Salmonella_enterica_enterica.fea.ods |
 |
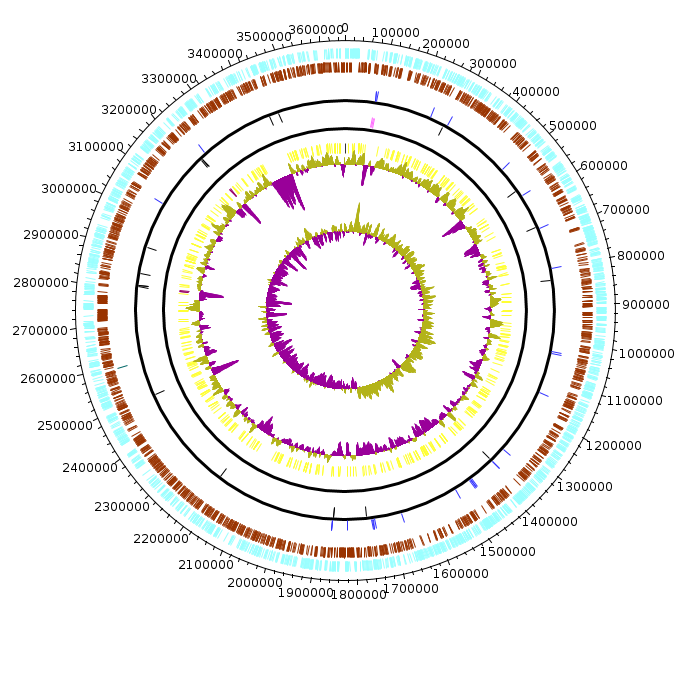
Sinorhizobium_meliloti_1021
Sinorhizobium_meliloti_1021.fea Sinorhizobium_meliloti_1021.fea.tsv Sinorhizobium_meliloti_1021.fea.ods |
 |
Streptococcus_mutans_LJ23
Streptococcus_mutans_LJ23.fea Streptococcus_mutans_LJ23.fea.tsv Streptococcus_mutans_LJ23.fea.ods |
 |